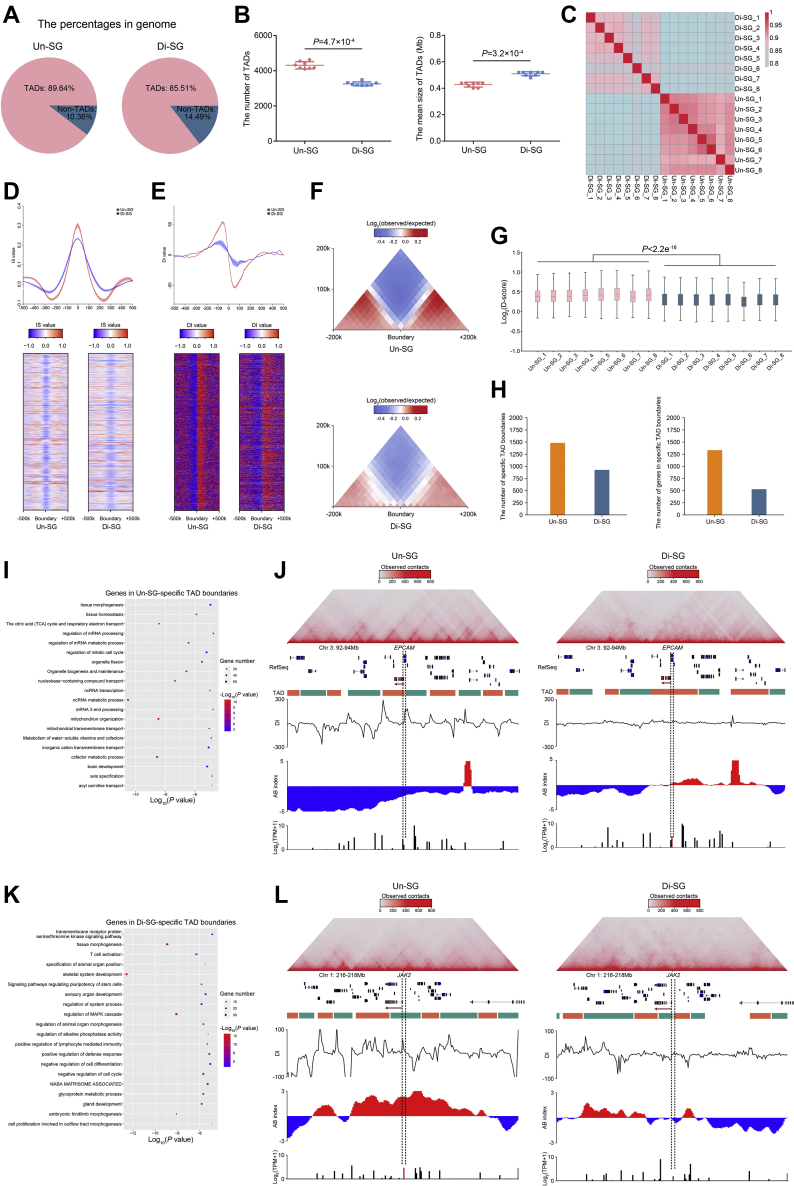
Figure 3.
Topologically associating domain dynamics during spermatogonial differentiation.A, the proportions of TADs and non-TADs in genome. B, the numbers (left) and mean sizes (right) of TADs in Un-SG and Di-SG. The data are presented as the mean ± SD of eight biological samples. P: Mann-Whitney U test, one-tailed. C, Jaccard indices illustrating the correlation of TAD architecture between Un-SG and Di-SG samples. D and E, the mean IS (D) and DI (E) value of TADs and the flanking regions (±500k) in Un-SG and Di-SG. F, the aggregate Hi-C map showing the average observed/expected chromatin interaction frequencies at TADs and the flanking regions (±200k) in Un-SG and Di-SG. G, the D-score in all Un-SG and Di-SG samples. P: Mann-Whitney U test, one-tailed. H, the numbers of specific TAD boundaries (left) and their harbored genes (right) in Un-SG and Di-SG. I, gene ontology-biological process analysis of genes in Un-SG-specific TAD boundaries. J, views of the observed/expected chromatin interaction frequencies (the upper panel), RefSeq (the middle panel), DI, A/B index, and RNA-seq coverage (the lower panel) at chromosome 3, 92 to 94 Mb, revealing that EPCAM was harbored in Un-SG-specific TAD boundaries, in B-A switching compartments and upregulated in Di-SG. K, gene ontology-biological process analysis of genes in Di-SG-specific TAD boundaries. L, views of the observed/expected chromatin interaction frequencies (the upper panel), RefSeq (the middle panel), DI, A/B index, and RNA-seq coverage (the lower panel) at chromosome 1, 216 to 218 Mb, revealing that JAK2 was harbored in Di-SG-specific TAD boundaries, in A-B switching compartments and downregulated in Di-SG. DI, directional index; Di-SG, differentiating spermatogonia; GO-BP,gene ontology-biological process; Hi-C, high-throughput chromosome conformation capture; IS, insulation score; TADs, topologically associating domains; Un-SG, undifferentiated spermatogonia.