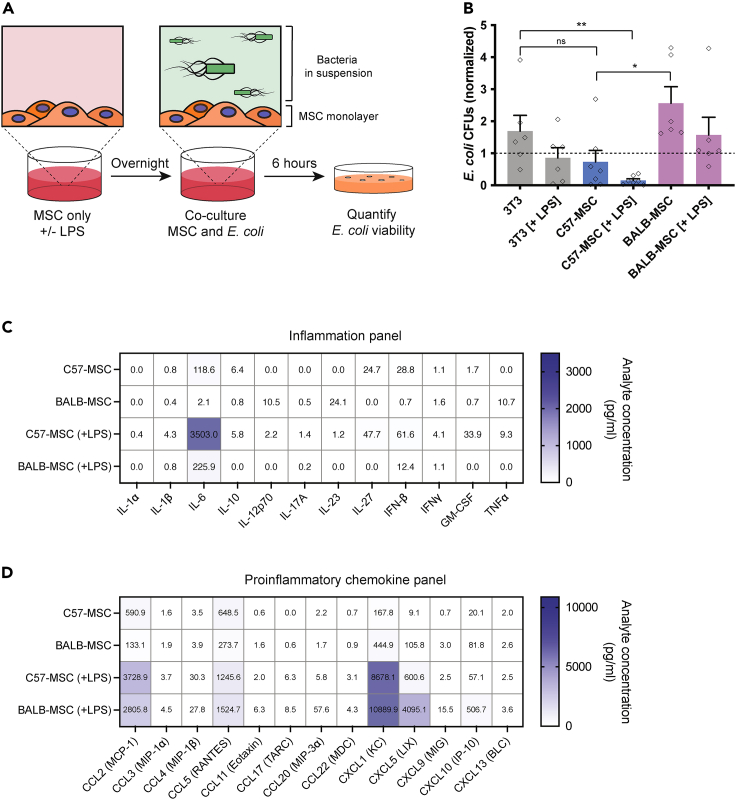
Figure 1.
MSCs from different genetic backgrounds have distinct antibacterial properties
(A) Cartoon depicting the antibacterial assays used in this study. MSCs were incubated overnight with or without lipopolysaccharide (LPS) from E. coli (100 ng/mL) and were then co-cultured with E. coli strain K12 for 6 h. After 6 h of co-culture, E. coli abundance in the media was quantified using colony-forming unit (CFU) assays.
(B) Quantification of E. coli CFUs following 6-h co-culture with different cell types +/− LPS treatment. E. coli CFU values from co-culture experiments were normalized to controls in which E. coli was grown side-by-side in monoculture (n = 5 biological replicates per condition; statistical significant determined using t test; ∗p < 0.05; ∗∗p < 0.01; individual data points are shown and bar graphs represent the mean with error bars = SEM).
(C and D) Cytokine profiling of C57-MSC and BALB-MSC conditioned media with or without LPS-priming using BioLegend LEGENDplex bead-based immunoassays with the mouse inflammation panel or (D) proinflammatory chemokine panel. Samples were analyzed 18-h post-LPS-exposure, and the values shown on heatmap are the mean of at least two biological replicates.