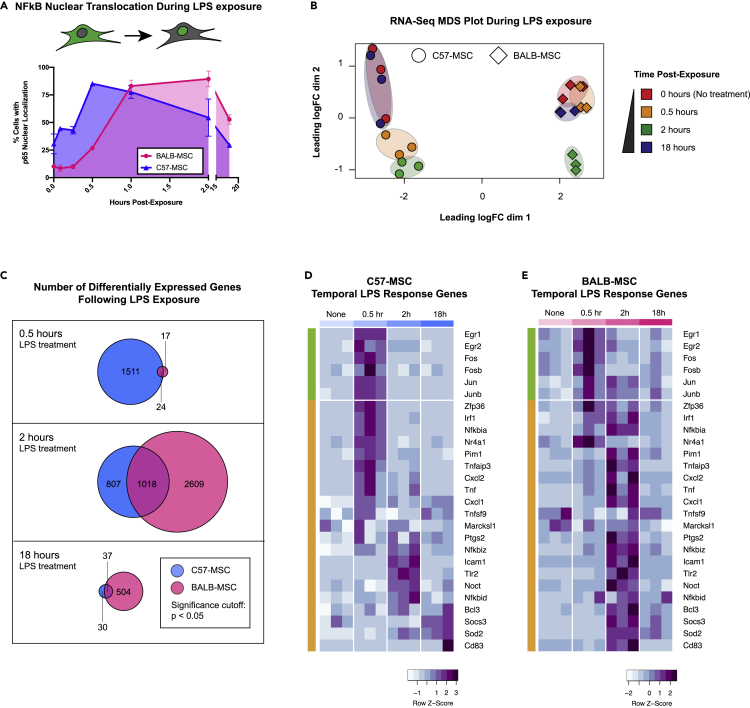
Figure 2.
Differential response rates to LPS between MSCs from different genetic backgrounds
(A) Quantification of NF-κB nuclear translocation assays using p65 staining and high-throughput imaging (>1000 cells analyzed per replicate; n = 3 biological replicates; error bars represent SD).
(B) MDS plot depicting transcriptional profiles of C57-MSCs and BALB-MSCs at time points post-LPS-exposure (0, 0.5, 2, and 18 h).
(C) Numbers of differently expressed genes at different time points post-LPS-exposure compared with corresponding untreated cells. (D and E) Heatmap depicting temporal expression of immediate early genes (green bars) and primary LPS response (orange bars) in C57-MSC and BALB-MSC at time points post-LPS-exposure (immediate-early genes and primary LPS response genes were previously defined (Bahrami and Drablos, 2016; Fowler et al., 2011; Ramirez-Carrozzi et al., 2009). For each time point analyzed, three biological replicates were sequenced and are depicted by sub-columns in the heatmap.