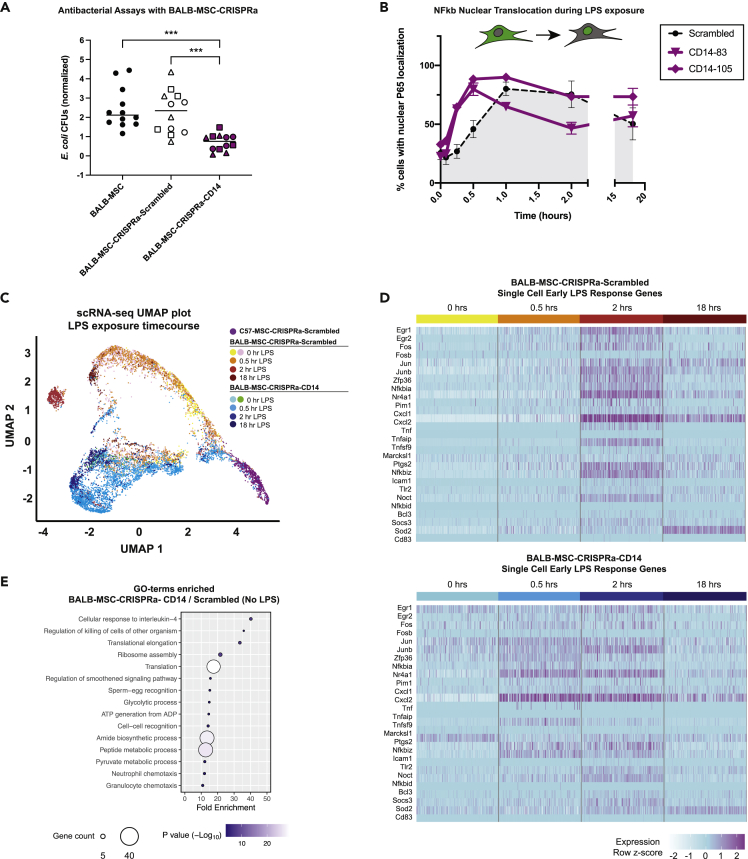
Figure 5.
Antibacterial and single-cell transcriptional analyses of BALB-MSCs overexpressing endogenous CD14 via CRISPRa
(A) E. coli CFUs after 6 h co-cultured with wild-type or BALB-MSC-CRISPRa cells. E. coli CFUs from co-cultures were normalized to E. coli monoculture CFU controls performed alongside each replicate. Three different sgRNAs were tested in CRISPRa MSCs, and the data from distinct sgRNAs are depicted by different shapes (for scrambled sgRNAs: circle = nontargeting sgRNA 1, square = nontargeting sgRNA 2, triangle = nontargeting sgRNA 3. For CD14 sgRNAs: circle = CD14-83, square = CD14-105, triangle = CD14-13). Lines are drawn at mean, and statistical significance was determined by t test; ∗∗∗, p < 0.001.
(B) Nuclear translocation of NF-κB during LPS-exposure using p65 immunostaining and quantitative microscopy in BALB-MSC-CRISPRa cells expressing a scrambled sgRNA or CD14 sgRNAs (scrambled sgRNA = nontargeting control one; n = at least three biological replicates per sample).
(C) UMAP plot depicting single-cell transcriptional profiles of MSCs during LPS-exposure (scrambled sgRNA = nontargeting control one; CD14 sgRNA = CD14-83). Single-cell RNA-seq of BALB-MSC overexpressing CD14 during LPS-exposure was prepared using the 10x Genomics Chromium platform followed by Illumina sequencing.
(D) Heatmap of early LPS response gene expression in 100 individual BALB-MSC-CRISPRa cells expressing either scrambled control sgRNA (top) or CD14 sgRNA (bottom). Each sub-column represents gene expression level in an individual cell.
(E) GO-terms enriched from significantly upregulated genes identified BALB-MSC-CRISPRa-CD14 when compared with BALB-MSC-CRISPRa-Scrambled in the absence of LPS-exposure.