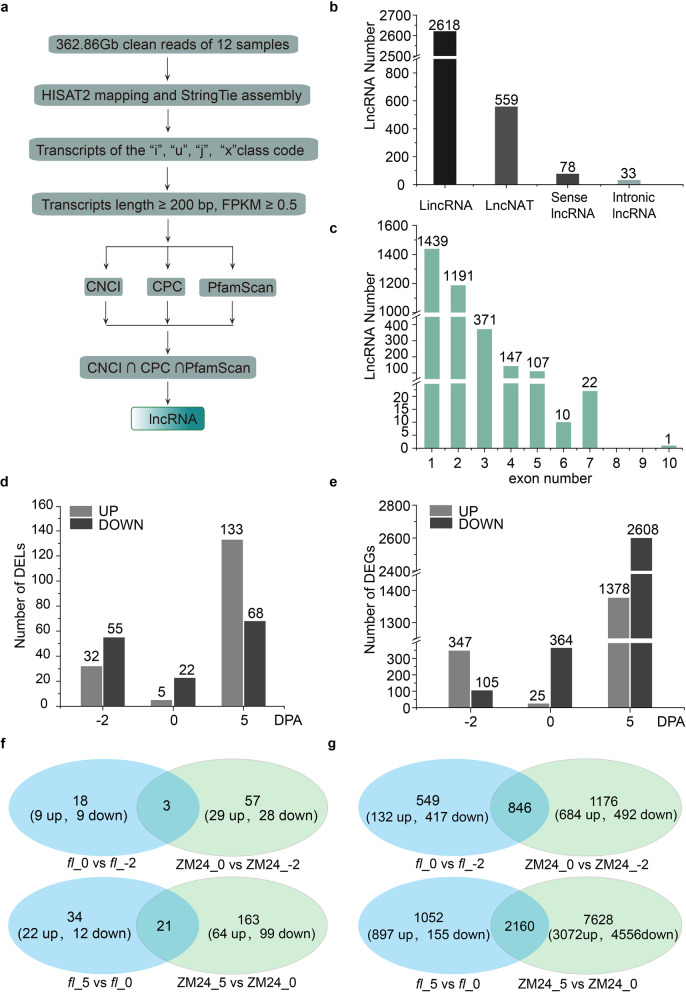
Fig. 2.
An integrative computational pipeline for the bioinformatics analysis and summary of DELs and DEGs in the RNA-Seq. a Bioinformatics methods for the identification of lncRNA. b The number of lincRNA, lncNAT, sense lncRNA, and ntronic lncRNA. c Exon number pattern of four kinds of lncRNA. d and e Number of differentially expressed lncRNAs and genes (up-and down-regulated) between fl and ZM24, respectively, in three time points (-2, 0, and 5 DPA) during fiber initiation. f and g Venn diagrams showing the common DELs and DEGs between comparisons of ‘0 DPA vs -2 DPA’ and ‘5 DPA vs 0 DPA’ of fl and ZM24, respectively. DELs and DEGs were identified with log2| (fold change)| > 1, FDR < 0.001