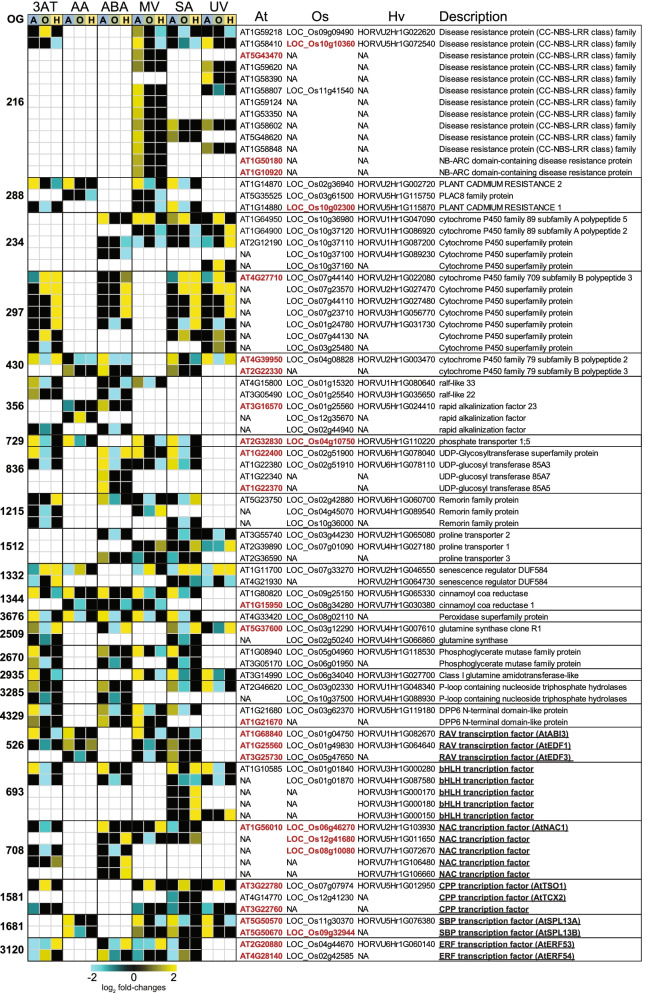
Fig. 6.
Oppositely responsive orthologous genes following treatments in Arabidopsis, rice and barley. Log2-transformed fold-changes (FDR < 0.05) are shown as a heatmap for the 24 orthogroups [24] that contained differentially expressed genes (DEGs) showing opposite responses (up/down-regulated) in 4 out of 6 treatments (3AT = 3-amino-1,2,4-triazole, AA = antimycin A, ABA = abscisic acid, MV = methyl viologen, SA = salicylic acid, UV = ultraviolet radiation) for each of the three species (A = Arabidopsis, O = Rice and H = Barley). Arabidopsis genes with known functions (ST 7b) in development, defence and/or redox homeostasis are indicated in red. Underlining indicates genes encoding transcription factors