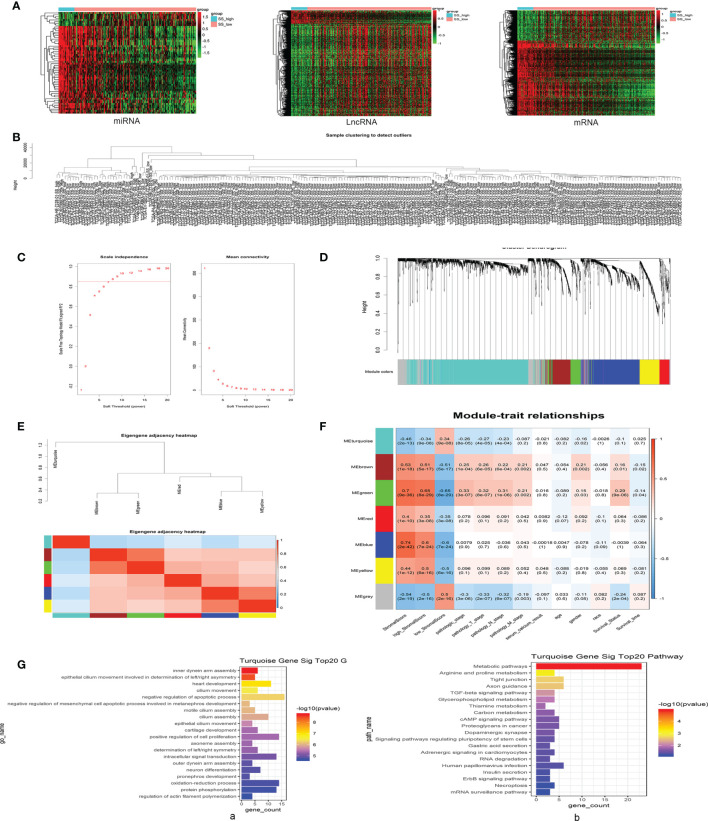
Figure 2.
Differentially expressed genes (DEGs) in patients with high vs. low SSs were evaluated using weighted gene co-expression network analysis (WGCNA), and the turquoise module was selected based on positive pathological roles in KIRP. (A) Unsupervised hierarchical clustering analyses for DEGs, including differentially expressed messenger (m)RNAs, long noncoding (lnc)RNAs, and micro (mi)RNAs, in SS-high and SS-low KIRP tissue samples. The cluster analysis heat map shows the correlation between expression maps and group conditions. The rows represent differentially expressed miRNAs, lncRNAs, and mRNAs, and the columns represent the samples. (B) Sample clustering detection revealed no outlier samples. (C) Soft-threshold power (β) for co-expression of lncRNAs/mRNAs was determined by analyzing the network topology with a soft-threshold power ranging from 1 to 20. (D) Different modules were identified by the Dynamic Tree Cutting method, and each module was assigned a color as an identifier. Six modules were generated after merging based on the correlation of modules with WGCNA. (E) Heatmap plot of the adjacencies in the hub gene network; red represents positive correlation with high adjacency, and blue represents negative correlation with low adjacency. Squares of red color along the diagonal represent the meta-module. (F) Matrix of module–trait relationships and P-values for selected traits. Each column corresponds to a module eigengene, and each row corresponds to a histopathological trait. Each cell contains a corresponding correlation and P-value. The table is color-coded by correlation according to the color legend. (G, a) Gene ontology (GO) enrichment analyses of turquoise mRNAs; significant top 20 GO terms are shown. (G, b) Kyoto Encyclopedia of Genes and genomes (KEGG)-pathway enrichment analyses of turquoise mRNAs; significant top 20 signaling pathways are shown.