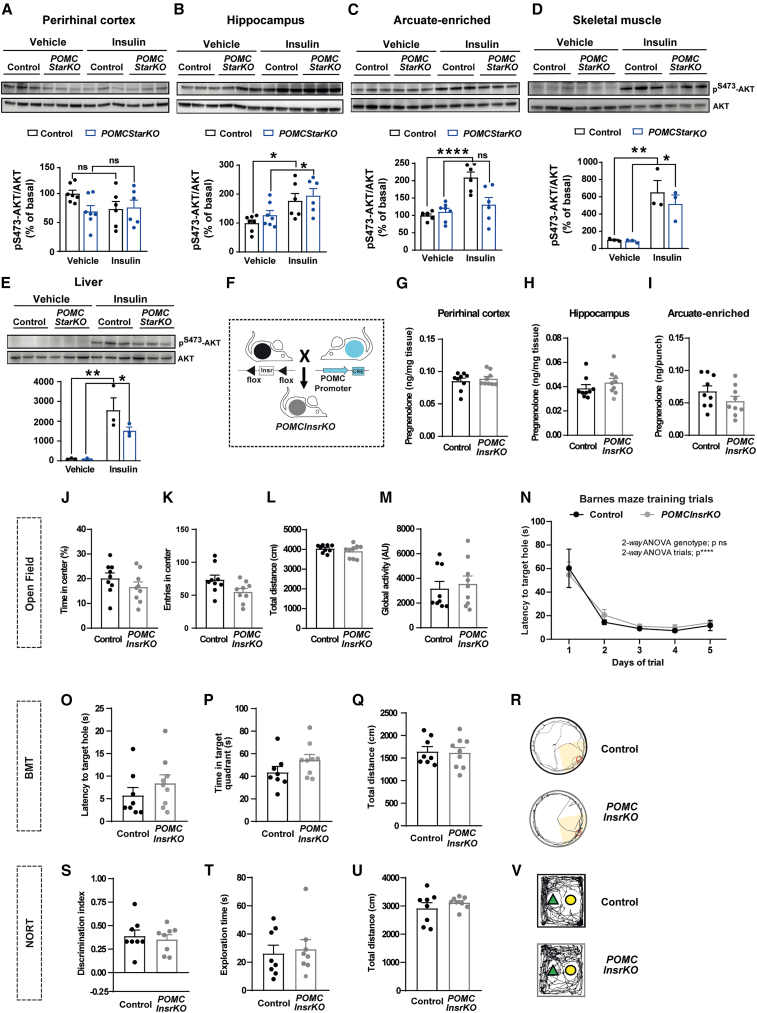
Figure 3.
Insulin signaling in POMC neurons does not mediate memory performance
(A–E) Assessment of insulin signaling via AKT phosphorylation (pS473-AKT) in the (A) perirhinal cortex, (B) hippocampus, (C) arcuate-enriched mediobasal hypothalamus, (D) skeletal muscle, and (E) liver from control and POMCStarKO mice. Representative blots and quantification (normalized to total AKT) are shown (n = 3–7/genotype).
(F) Schematic illustration of the generation of POMCInsrKO mice.
(G–I) Pregnenolone content in the (G) perirhinal cortex, (H) hippocampus, and (I) arcuate nucleus-enriched mediobasal hypothalamus from control and POMCInsrKO mice (n = 9/genotype).
(J–M) Locomotor and exploratory activity of control and POMCInsrKO mice in an open-field paradigm: (J) time spent in the center, (K) entries in the center, (L) total distance traveled, and (M) global activity (n = 9/genotype).
(N) Task learning curve for the latency to reach the scape hole during the training phase of the Barnes maze test (BMT). Results show the average of 2 trials per day during 5 consecutive days (n = 8–9/genotype).
(O–R) Recorded parameters to assess Barnes maze performance in control and POMCInsrKO mice during the test phase: (O) latency to target hole, (P) time spent in target quadrant, (Q) total distance traveled, and (R) representative traces of mouse movement trajectories (n = 8–9/genotype).
(S–V) Recorded parameters to assess NORT performance in control and POMCInsrKO mice during the test phase: (S) discrimination index (time exploring novel object − time exploring familiar object)/(time exploring novel object + time exploring familiar object), (T) exploration time (time exploring novel object + time exploring familiar object), (U) total distance traveled, and (V) representative traces of mouse movement trajectories (n = 8/genotype).
All studies were performed in male mice between 10 and 16 weeks of age. Dots in panels represent individual samples. Data are presented as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001; ns, not significant.