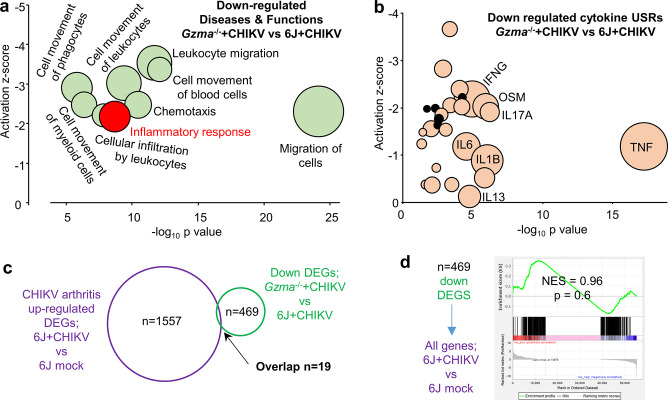
Figure 2. RNA-Seq of Gzma-/- + chikungunya virus (CHIKV) vs. 6J + CHIKV day 6 feet.
(a) Selected Ingenuity Pathway Analysis (IPA) Diseases and Function annotations for the 469 downregulated differentially expressed genes (DEGs) for Gzma-/- + CHIKV vs. 6J + CHIKV (full set of annotations shown in Supplementary file 2c). (b) IPA cytokine upstream regulators (USRs) downregulated in CHIKV-infected feet of Gzma-/- mice (Gzma-/-+CHIKV vs. 6J + CHIKV; Supplementary file 2e) plotted by p-value and z-score. Black circles – minor USRs for Gzma-/- + CHIKV vs. 6J + CHIKV not identified for 6J + CHIKV vs. 6J mock (see also Figure 4e). (c) RNA-Seq identified 1557 DEGs upregulated in feet for 6J + CHIKV vs. 6J mock infection (Supplementary file 2g). RNA-Seq of Gzma-/- + CHIKV vs. 6J + CHIKV day 6 feet identified 469 downregulated DEGs in Gzma-/- mice associated with the reduced foot swelling (Supplementary file 2b). Only 19 of these DEGs were shared by these datasets. (d) Gene Set Enrichment Analysis (GSEA) of downregulated DEGs from Gzma-/- + CHIKV vs. 6J + CHIKV day 6 feet (Supplementary file 2b) vs. all genes (preranked by fold change) from feet 6J + CHIKV vs. 6J mock infection (Supplementary file 2f).