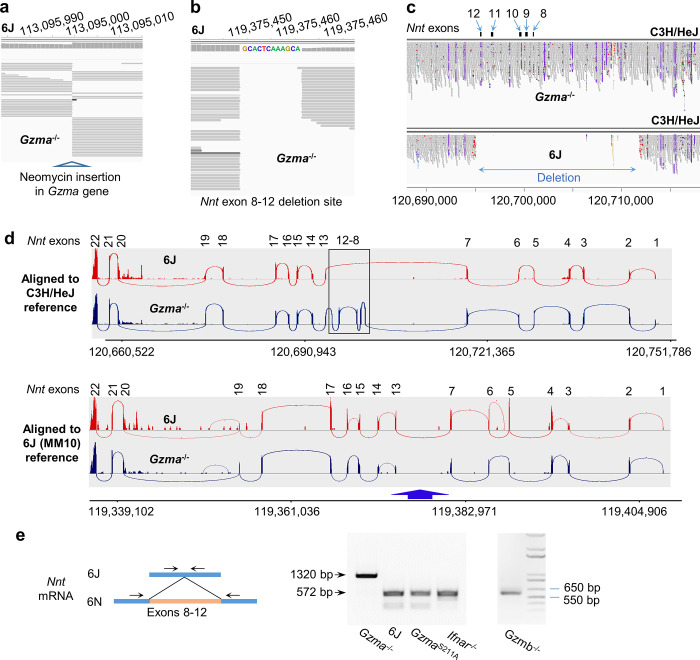
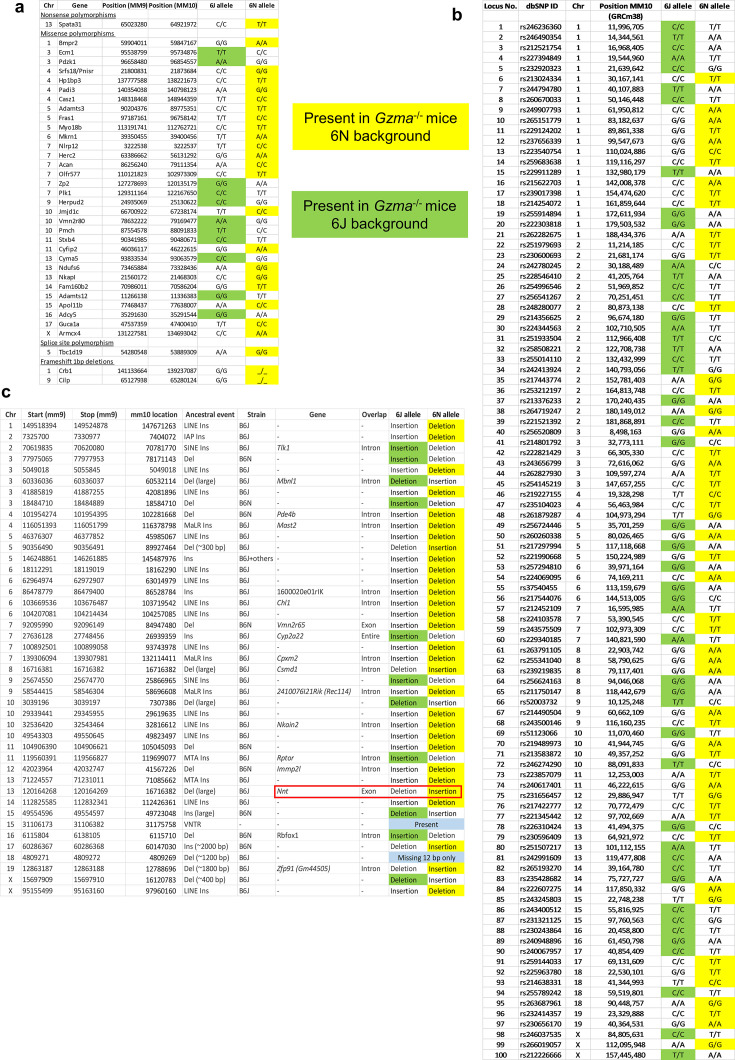
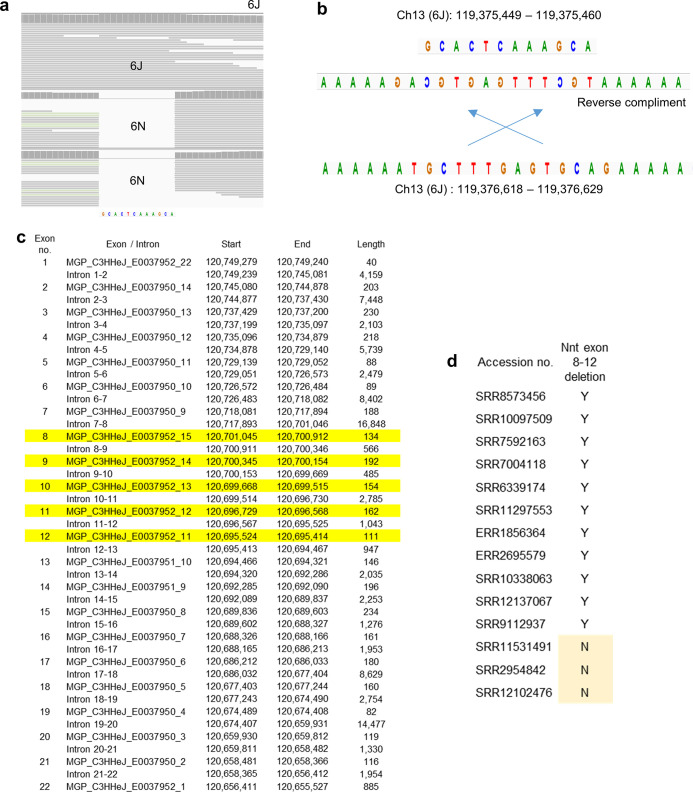
Figure 3. Nnt deletion in 6J but not Gzma-/- mice.
(a) Whole-genome sequencing (WGS) of Gzma-/- mice aligned to the 6J (MM10) reference genome build, illustrating the insertion site of the neomycin cassette into the Gzma gene to create the knockout. (b) As for (a) but showing the site of the Nnt deletion, with the additional 12 nucleotides present in the 6J genome. (c) WGS of Gzma-/- and 6J mice aligned to the C3H/HeJ reference genome, illustrating that the Nnt deletion present in 6J mice is absent in Gzma-/- mice. The deletion is in chromosome 13; position 120,695,141–120,711,874 (C3H/HeJ numbering). (d) Reads from RNA-Seq of chikungunya virus (CHIKV)-infected 6J and Gzma-/- mice aligned to the C3H/HeJ and 6J (MM10) reference genomes showing the Sashimi plot (Integrative Genomics Viewer) for the Nnt gene. (e) RT-PCR of testes using primers either side of exons 8–12 in the Nnt mRNA.