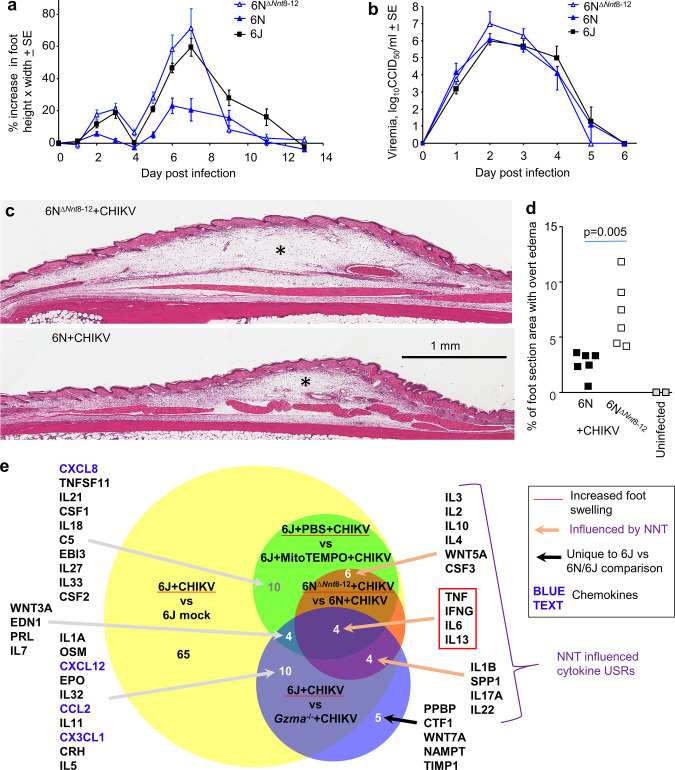
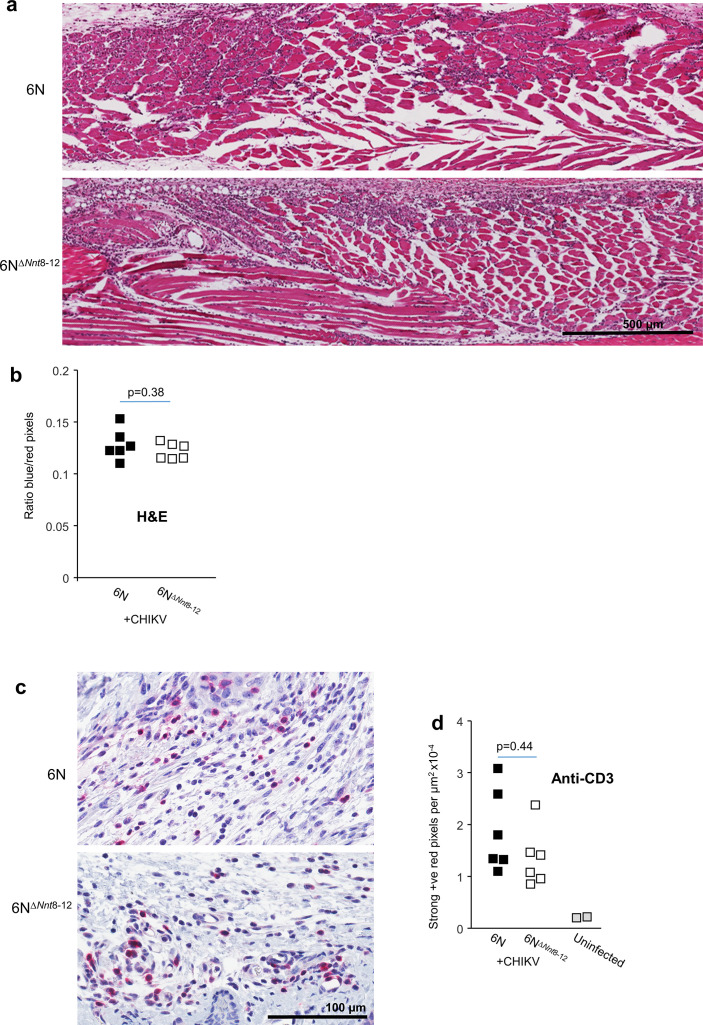
Figure 4. 6N∆Nnt8-12 mice.
(a) 6N∆Nnt8-12 mice have the same Nnt exon deletion as 6J mice. Age-matched female 6N∆Nnt8-12, 6N, and 6J mice were infected with chikungunya virus (CHIKV) and foot swelling measured over time (n = 5 mice and 10 feet per group). Foot swelling was significantly higher in 6N∆Nnt8-12 mice when compared with 6N mice on days 2–7 (day 2 p=0.0026, day 7 p=0.0027, t-tests, parametric data distributions; days 3–6 p=0.003, Kolmogorov–Smirnov tests, nonparametric data distributions). Foot swelling was significantly lower in 6N mice when compared with 6J mice (day 2 p=0.042, day 6 p=0.001, day 7 p=0.0005, t-tests, parametric data distributions; days 3 and 5, p=0.002, Kolmogorov–Smirnov tests, nonparametric data distributions). (b) Viremia for the same mice as in (a). (c) H&E staining of feet from 6N∆Nnt8-12 and 6N mice day 6 post infection showing subcutaneous edema (*). (d) Percentage of foot section area showing overt subcutaneous edema (statistics by Kolmogorov–Smirnov test). (e) RNA-Seq data for four comparisons was analyzed by Ingenuity Pathway Analysis (IPA) and cytokine upstream regulator (USR) overlaps shown. Only cytokine USRs with positive z-scores associated with increased foot swelling are shown.