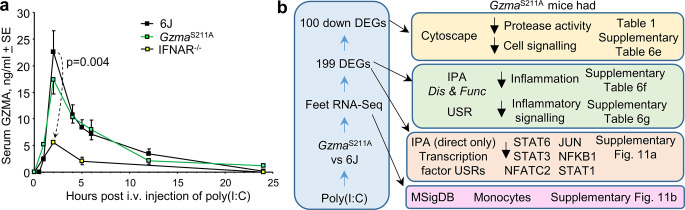
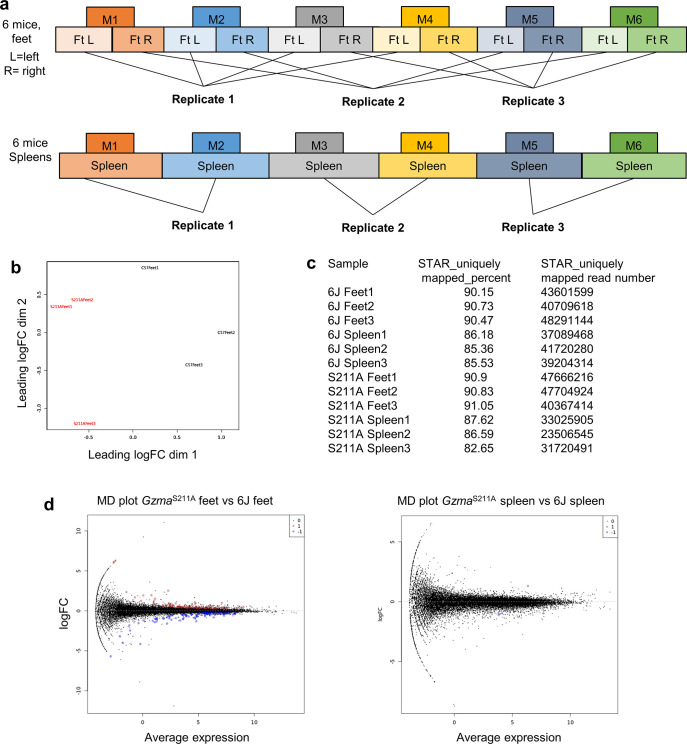
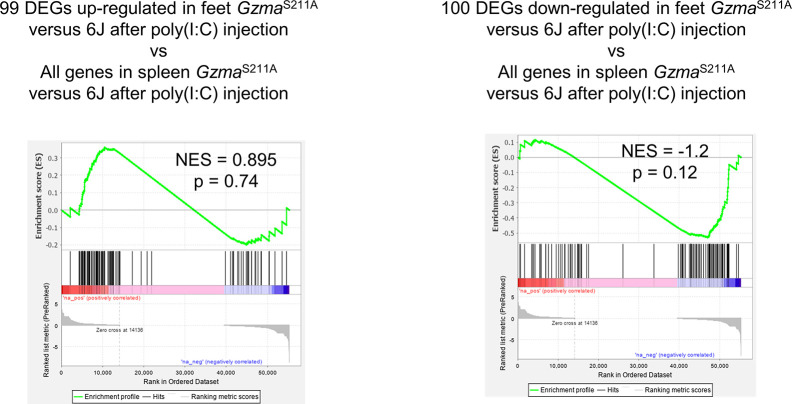
Figure 6. Polyinosinic:polycytidylic acid (poly(I:C)) injection into GzmaS211A, 6J mice, and Ifnar-/-.
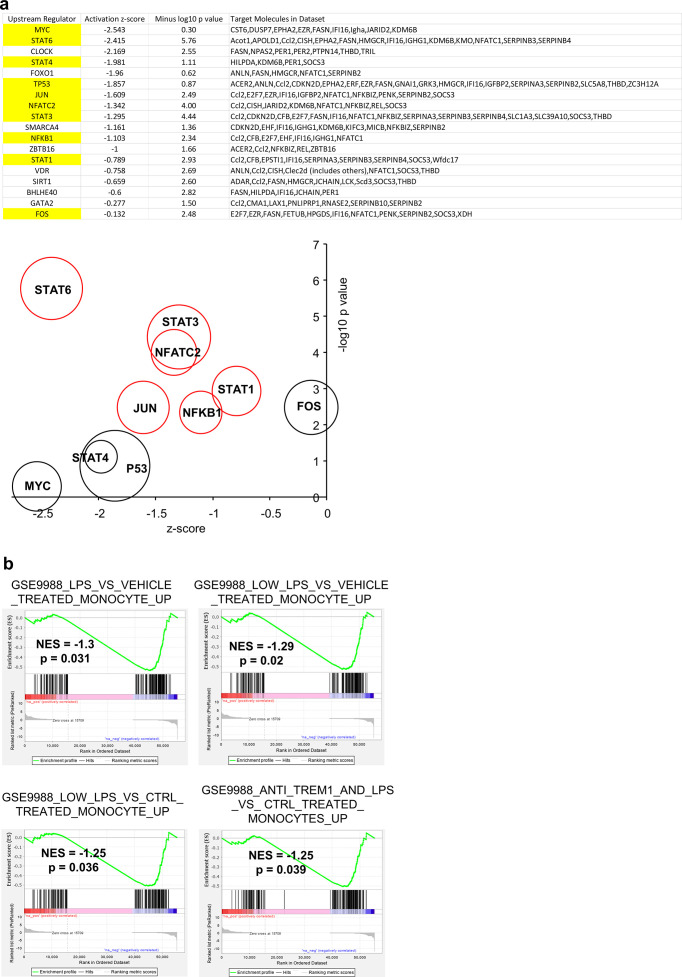
(a) GzmaS211A, 6J, and Ifnar-/- mice were injected i.v. with 250 µg of poly(I:C) in 150 µl of PBS, and serum samples were taken at the indicated times and assayed for GZMA concentration using a capture ELISA kit (6J n = 5–8, Ifnar-/- n = 5–6 and GzmaS211A n = 3 mice per time point). (b) GzmaS211A and 6J mice were injected i.v. with 250 µg of poly(I:C) and feet removed 6 hr later and analyzed by RNA-Seq. The differentially expressed genes (DEGs) (Supplementary file 6b) were analyzed by Cytoscape and Ingenuity Pathway Analysis (IPA). The full gene list (Supplementary file 6a) was analyzed using the Molecular Signature Database (MSigDB).