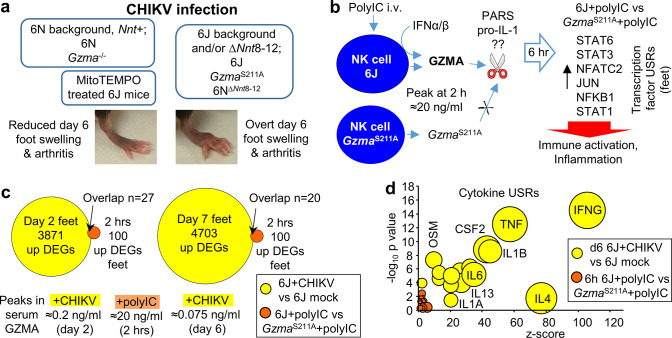
Figure 7. Summary of chikungunya virus (CHIKV) foot swelling and granzyme A (GZMA) bioactivity, and the poor concordance between the two.
(a) Summary of foot swelling results. 6N and Gzma-/- mice have a 6N or mixed 6J/6N background and have an intact Nnt gene and show reduced CHIKV-induced foot swelling. MitoTEMPO treatment also reduced foot swelling. 6J, GzmaS211A, and 6N∆Nnt8-12 mice are all missing exons 8–12 of Nnt and show increased foot swelling. (b) Summary of GZMA bioactivity. Using polyinosinic:polycytidylic acid (poly(I:C)) to induce high levels of GZMA secretion from NK cells, studies in GzmaS211A mice illustrated that proteolytically active circulating GZMA promotes certain immune-stimulating/pro-inflammatory responses (dominant transcription factor upstream regulators (USRs) are shown; Figure S11a). No clear consensus has emerged regarding the molecular target(s) of GZMA (??); two potential extracellular candidate targets are shown; protease activated receptors and pro-IL-1. (c) Low overlap between CHIKV and GZMA induced differentially expressed genes (DEGs). DEGs upregulated in feet by CHIKV infection of 6J mice on days 2 and 7 (a low stringency filter of q < 0.05 was applied to the all gene lists in Supplementary file 2f to provide these DEGs) were compared with the DEGs upregulated in feet by active GZMA in 6J mice 6 hr after poly(I:C) treatment (i.e., downregulated in GzmaS211A mice; Supplementary file 6b). Overlapping genes (n = 27 and 20) are listed in Supplementary file 6i. The mean peak levels of serum GZMA for each group are shown below the Venn diagrams. (d) Cytokine USRs for GZMA vs. CHIKV. A series of cytokine USRs were induced by proteolytically active GZMA (i.e., upregulated for 6J + polyIC vs. GzmaS211A + polyIC; Supplementary file 6g, column R). The same USRs were significantly more upregulated by CHIKV infection (Supplementary file 2i). Data for GZMA (orange) and CHIKV (yellow) are plotted with bubble size representing number of molecules in dataset.