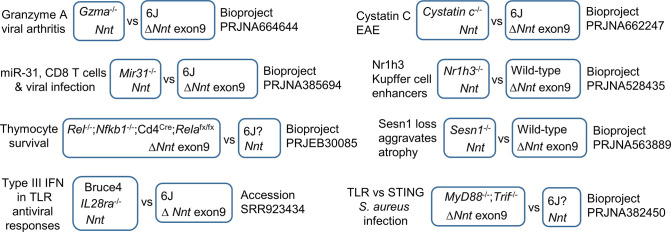
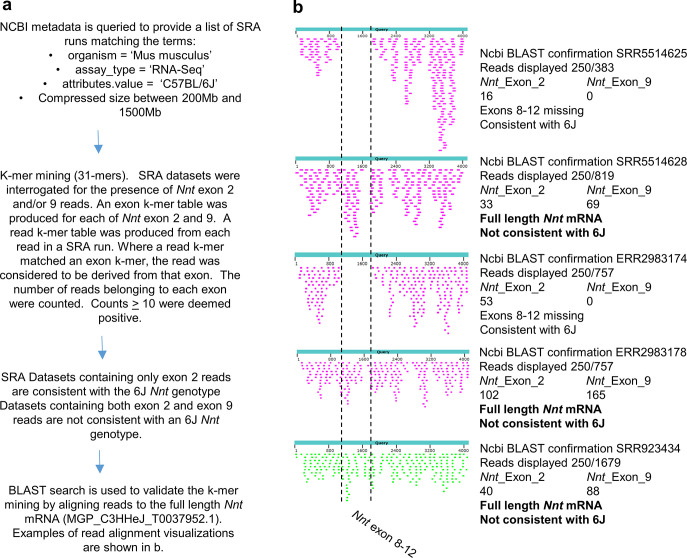
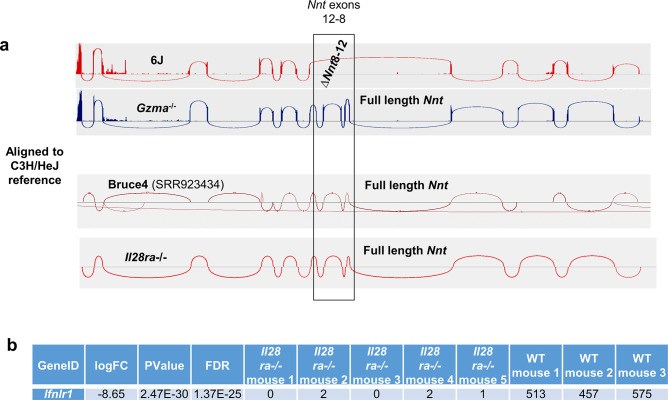
Figure 8. k-mer mining of BioProjects where Nnt+ mice were compared with Nnt- mice.
The NCBI Sequence Read Archive (SRA) database was interrogated by k-mer mining for BioProjects where (i) some Run Accessions (listing 6J as the mouse strain) had reads compatible with a 6J background (reads for Nnt exon 2, but not exon 9) and (ii) other Run Accessions in that BioProject (listing 6J as the mouse strain) had reads not compatible with a 6J background (reads for Nnt exons 2 and 9). The methodology is described in Figure 8—figure supplement 1a, validated by BLAST alignments (Figure 8—figure supplement 1b), with raw data in Supplementary file 7d and e.