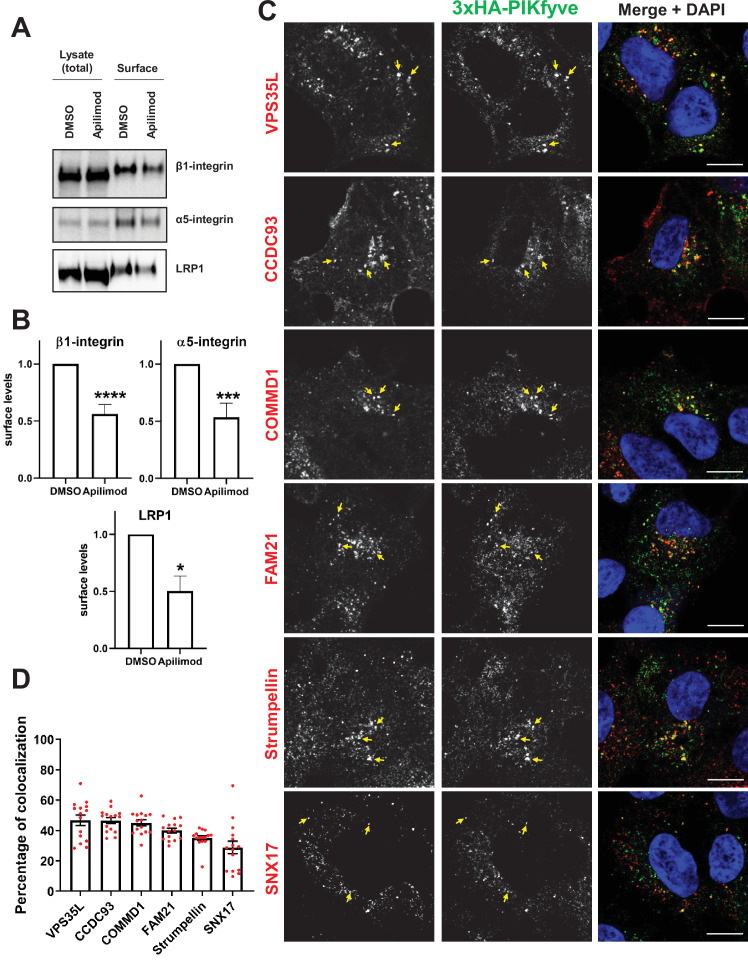
Figure 6. PIKfyve regulates the recycling of SNX17-Retriever-CCC-WASH cargoes and colocalizes with SNX17 and subunits of the Retriever, CCC, and WASH complexes.
(A) HeLa cells were treated with DMSO or 1 µM apilimod for 1 hr and then the levels of SNX17 cargoes, β1-integrin, and α5-integrin were determined using a surface biotinylation assay. Surface biotinylation assay was similarly performed on HUH7 cells and low-density lipoprotein receptor-related protein 1 (LRP1) levels were measured. (B) Quantification of western blots from three independent surface biotinylation experiments. (C) HEK293 cells expressing 3xHA-endogenously tagged PIKfyve were fixed, permeabilized, and incubated with antibodies against the HA tag and antibodies against either SNX17, the Retriever complex subunit (VPS35L), CCC complex subunits (CCDC93 and COMMD1), or WASH complex subunits (FAM21 and Strumpellin). Bar: 10 µm. Arrows indicate examples of puncta showing colocalization. (D) The percentage of PIKfyve colocalizing with the indicated proteins was determined using Mander’s colocalization coefficient analysis from three independent experiments. Data presented as mean ± SE. *p < 0.05, ***p < 0.005, and ****p < 0.001.
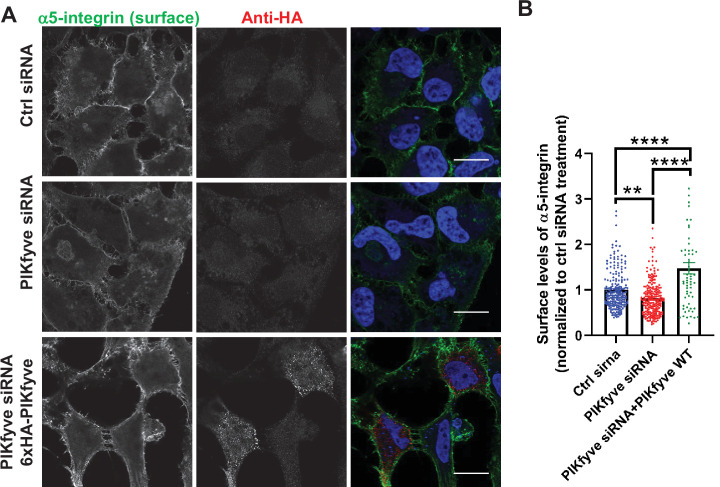
Figure 6—figure supplement 1. Depletion of PIKfyve results in a decrease of the surface levels of α5-integrins.
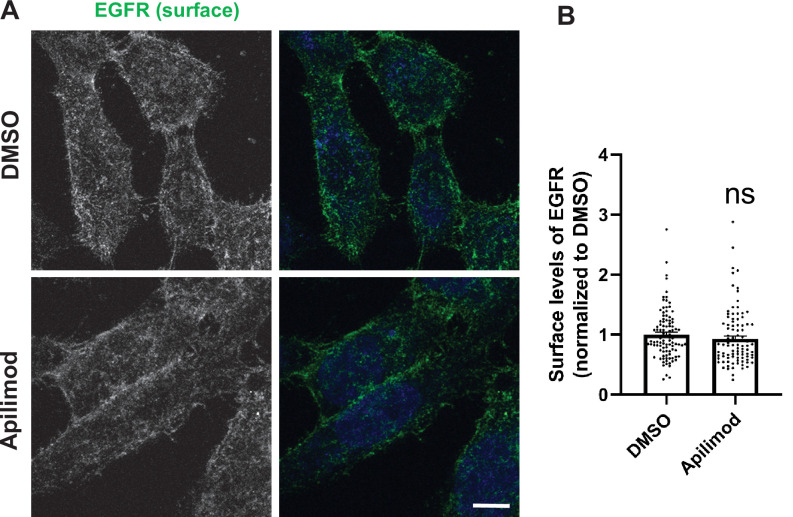
Figure 6—figure supplement 2. Inhibition of PIKfyve does not alter the surface levels of EGFR.
Figure 6—figure supplement 3. Immunofluorescence localization of endosomal proteins in unedited HEK293 cells (control for Figure 6).