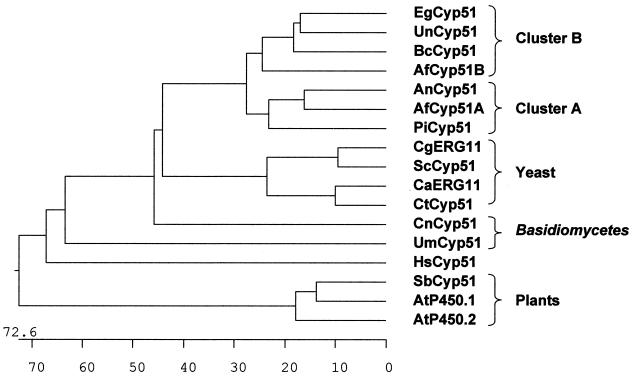
FIG. 5.
Phylogenetic tree derived by CLUSTAL analysis after the alignment of the full amino acid sequences of Cyp51A and Cyp51B from A. fumigatus with CYP51s from C. albicans (CaErg11; GenBank accession no. AAF00598.1), C. glabrata (CgERG11; GenBank accession no. AAB02329.1), C. tropicalis (CtCyp51; GenBank accession no. AAA53284.1), S. cerevisiae (ScCyp51; GenBank accession no. AAA34546.1), P. italicum (PiCyp51; GenBank accession no. CAA89824.1), A. nidulans (AnCyp51; GenBank accession no. AAF79204.1), E. graminis (EgCyp51; GenBank accession no. AAC97606.1), U. necator (UnCyp51; GenBank accession no. AAC49812.2), B. cinerea (BcCyp51; GenBank accession no. AAF85983.1), U. maydis (UmCyp51; GenBank accession no. CAA88176.1), C. neoformans (CnCyp51; GenBank accession no. AAF35366.1), A. thaliana (AtP450.1 [GenBank accession no. AAD.30262.1] and AtP450.2 [GenBank accession no. AAB86510.1]), S. bicolor (SbCyp51; GenBank accession no. AAC49659.1), and H. sapiens (HsCyp51; GenBank accession no. XP004663.1). The length of each branch represents the distance between sequence pairs. Numbers at the bottom of the tree are numbers of substitution events.