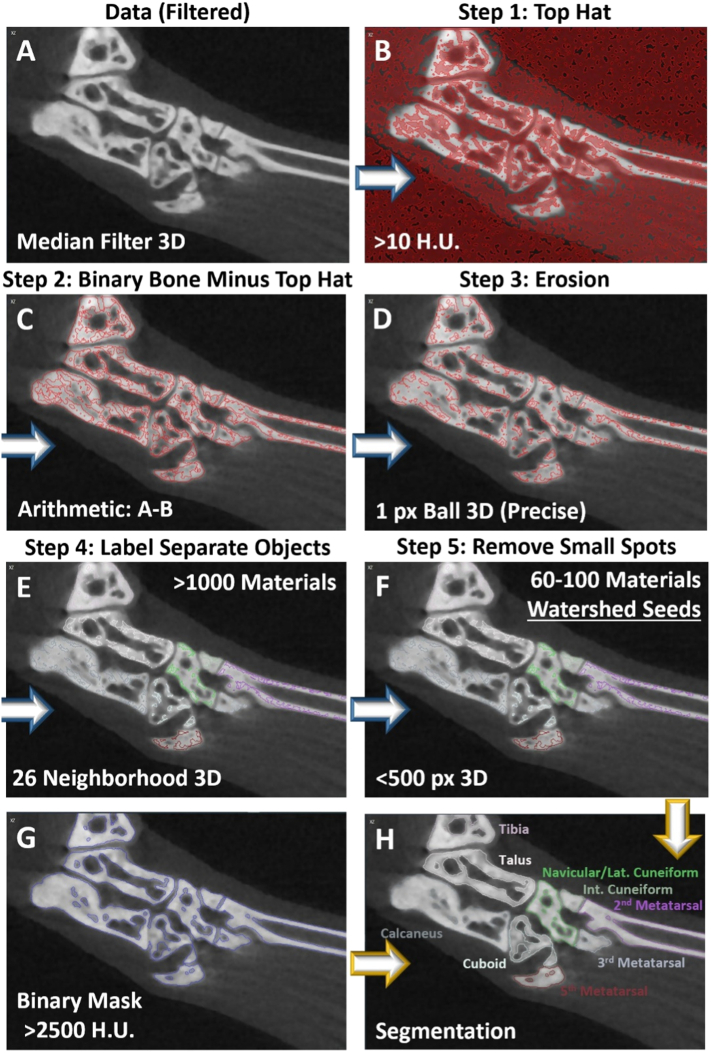
Fig. 3.
Automated workflow for generation of bone-specific watershed seeds. To create the bone-specific watershed seeds in an efficient and reproducible manner, an automated workflow was developed and packaged as a convenient “Recipe” where all steps are embedded within a single module in Amira. To visualize the changes at each step in the recipe, a 2D section near the tarsal region of a representative hindpaw is shown, while the modules function on the full 3D dataset. The recipe requires the input of the dataset after application of a three-dimensional median filter (A) and a binary representation of the bone (>2500 HU). A top-hat was then applied to identify local valleys in signal intensity, and a threshold >10 HU was set to define the valley depth (B). The top hat segmentation was then subtracted from the binary representation of the bone to define only high-intensity regions with limited signal change in adjacent voxels (C). The result was then eroded to further separate the intense regions of the individual bones (D), and the separate objects were then defined as individual materials with connected voxels sharing at least one common vertex (E). Small objects were then removed to isolate the larger connected materials defining the majority of the individual bones as the final step in the generation of the watershed seeds (F). An automated workflow was used to create the binary mask, which utilizes the same approach as shown in A-C, but with an increased threshold for valley depth at >750 HU in the top-hat step (G). The filtered dataset (A), watershed seeds (bone-specific, color-annotated contours) (F), and binary mask (blue contours) (G) were combined together in the “Marker Based Watershed Inside Mask” Amira module for the complete segmentation of the individual bones in the hindpaw (H), as described in Figure 2.