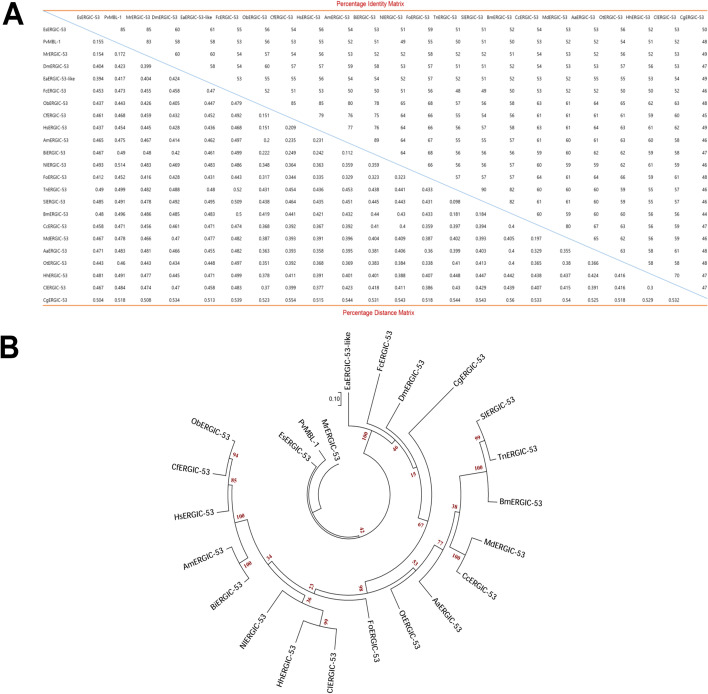
Fig. 5.
Alignment of MrERGIC-53 with orthologs from representative species. (A) Percentage identity and distance matrix of MrERGIC-53 and representative ERGIC-53 protein sequences from representative species. The analysis was performed using ClustalX2 (Version 2.0). The percentage identity matrix is shown from left to right, and the percentage distance matrix is shown from top to bottom. (B) Phylogenetic analysis of MrERGIC-53 with ERGIC-53 protein sequences from representative species. The consensus maximum likelihood tree (Tree Explorer, circular original tree) was built on the basis of the JTT matrix-based model using MEGA (Version 7.0) software. The bootstrap values are indicated by the numbers at the nodes. The ERGIC-53 sequences were selected from the following representative species: Eriocheir sinensis (EsERGIC-53, AIM45537.1), Penaeus vannamei (PvMBL-1, ROT63057.1), Daphnia magna (DmERGIC-53, JAN85016.1), Eurytemora affinis (EaERGIC-53-like, XP_023340026.1), Folsomia candida (FaERGIC-53, OXA55457.1), Ooceraea biroi (ObERGIC-53, XP_011335262.2), Camponotus floridanus (CfERGIC-53), Harpegnathos saltator (HsERGIC-53, XP_011154813.1), Apis mellifera (AmERGIC-53, XP_006565093.1), Bombus impatiens (BiERGIC-53, XP_012246683.1), Nilaparvata lugens (NlERGIC-53, XP_022197547.1), Frankliniella occidentalis (FoERGIC-53, XP_026275094.1), Trichoplusia ni (TnERGIC-53, XP_02673666.1), Spodoptera litura (SlERGIC-53, XP_022817189.1), Bombyx mori (BmERGIC-53, XP_012546331.1), Ceratitis capitata (CcERGIC-53, XP_004525276.2), Musca domestica (MdERGIC-53, XP_005179276.1), Aedes aegypti (AaERGIC-53, XP_021697592.1), Onthophagus taurus (OtERGIC-53, XP_022911466.1), Halyomorpha halys (HhERGIC-53, XP_014294482.1), Cimex lectularius (ClERGIC-53, XP_014245666.1), and Crassostrea gigas (CgERGIC-53, XP_011441168.1)