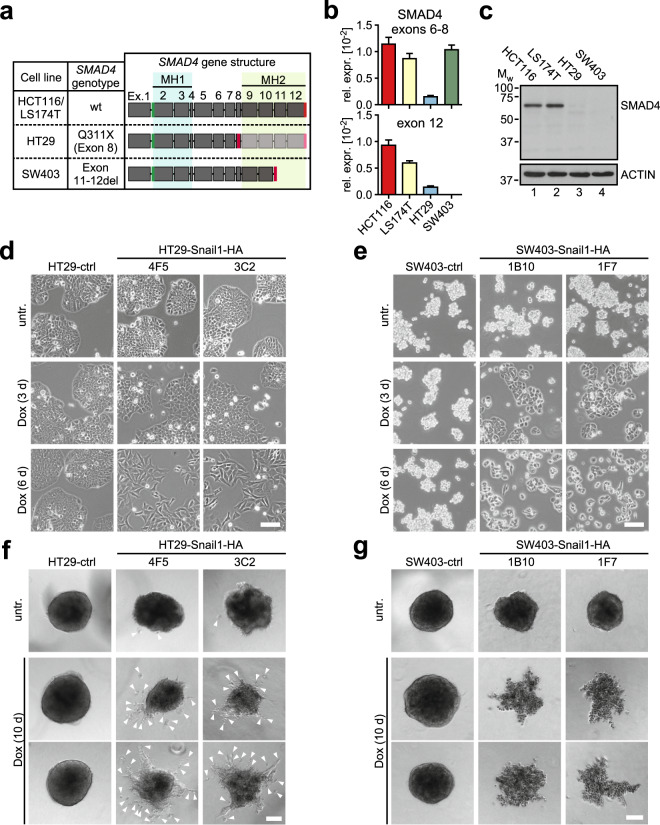
Fig. 1. HT29 and SW403 CRC cell lines are SMAD4-negative but show morphological evidence of EMT upon Snail1-HA overexpression.
a SMAD4 mutation status in four CRC cell lines and schematic depictions of respective gene structures and their coding capacities. Exon numbers as well as locations of the two major functional domains MH1 and MH2 are given on top. Positions of start codons are marked in green, stop codons are indicated in red. Introns are not drawn to scale. The precise location of the stop codon in SW403 cells is not known. b Analyses of SMAD4 mRNA expression by qRT-PCR in the four CRC cell lines. Primers were located in exons 6 and 8, and in exon 12. Relative gene expression (rel. expr.) was calculated by normalizing to the expression of GAPDH. Plotted is the mean + SEM; n = 3. c Analyses of SMAD4 protein expression in the four CRC cell lines by immunoblotting. Positions of molecular weight (Mw) standards in kDa are indicated on the left. ACTIN was detected as a loading control. d Representative phase contrast images of HT29-ctrl cells and HT29-Snail1-HA clones 4F5 and 3C2. Cells were left untreated (untr.) or received Dox for the indicated time spans. Scale bar: 100 µm. e Representative phase contrast images of SW403-ctrl cells and SW403-Snail1-HA clones 1B10 and 1F7. Cells were left untreated or received Dox for the indicated time spans. Scale bar: 100 µm. f Representative spheroids formed by HT29-ctrl cells and HT29-Snail1-HA clones 4F5 and 3C2 in a collagen I matrix after treatment with Dox as indicated. Cells that are separated from the spheroid bodies are highlighted by white arrowheads and were counted for quantification of cell invasiveness (see Fig. S2e). Scale bar: 100 µm. g Representative spheroids formed by SW403-ctrl cells and SW403-Snail1-HA clones 1B10 and 1F7 in a collagen I matrix after treatment with Dox as indicated. Scale bar: 100 µm.