Figure 3. Molecular design of the mammalian daily clock.
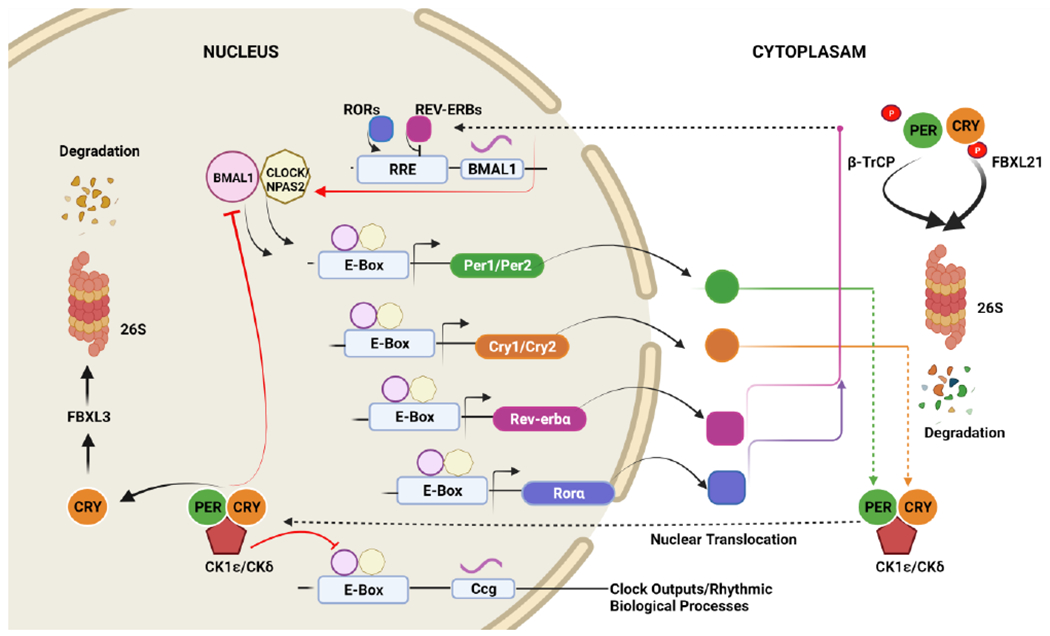
The core molecular clock is comprised of positive regulators (Bmal1 and CLOCK) and negative regulators (cryptochrome (Cry 1/2) and period-Per1/2). The protein clock Bmal1 heterodimerizes with CLOCK/NPAS2 to initiate the transcription of target genes through E-boxes present in the promoter region of target genes, resulting in the expression of Per (green circles) and Cry (orange circles) genes. Per and Cry dimerizes and translocates to the nucleus after being phosphorylated by casein kinase 1 epsilon (CK1ε) and casein kinase 1 delta (CK1δ), respectively, and repress their transcription. The Per and Cry proteins inhibit the expression of Bmal1 and CLOCK, creating the core negative feedback loop. In turn, CLOCK-Bmal1 generates another negative feedback loop that upregulates the negative transcription of REV-ERB proteins and negatively regulates Bmal1 transcription, and RORα binding to RORE elements increases the expression of Bmal1. Additionally, Per and Cry genes are regulated by proteins such as AMP-activated protein kinase (AMPK) and CK1ε/δ by phosphorylation and subsequent degradation. This phosphorylation targets Per genesfor polyubiquitination by βTrCP and, similarly, Cry1/Cry2 are phosphorylated by the Fox- proteins, FBXL3 and FBXL21, directing them to ubiquitin-mediated proteasomal degradation. Collectively, these feedback loops trigger circadian oscillations and subsequently regulate the circadian expression of downstream clock-controlled genes (CCGs). (Created with BioRender.com)
