Figure 1. Integrated modeling of post-operative day 1 (POD1) immune responses categorizes patients with and without a surgical site complication (SSC).
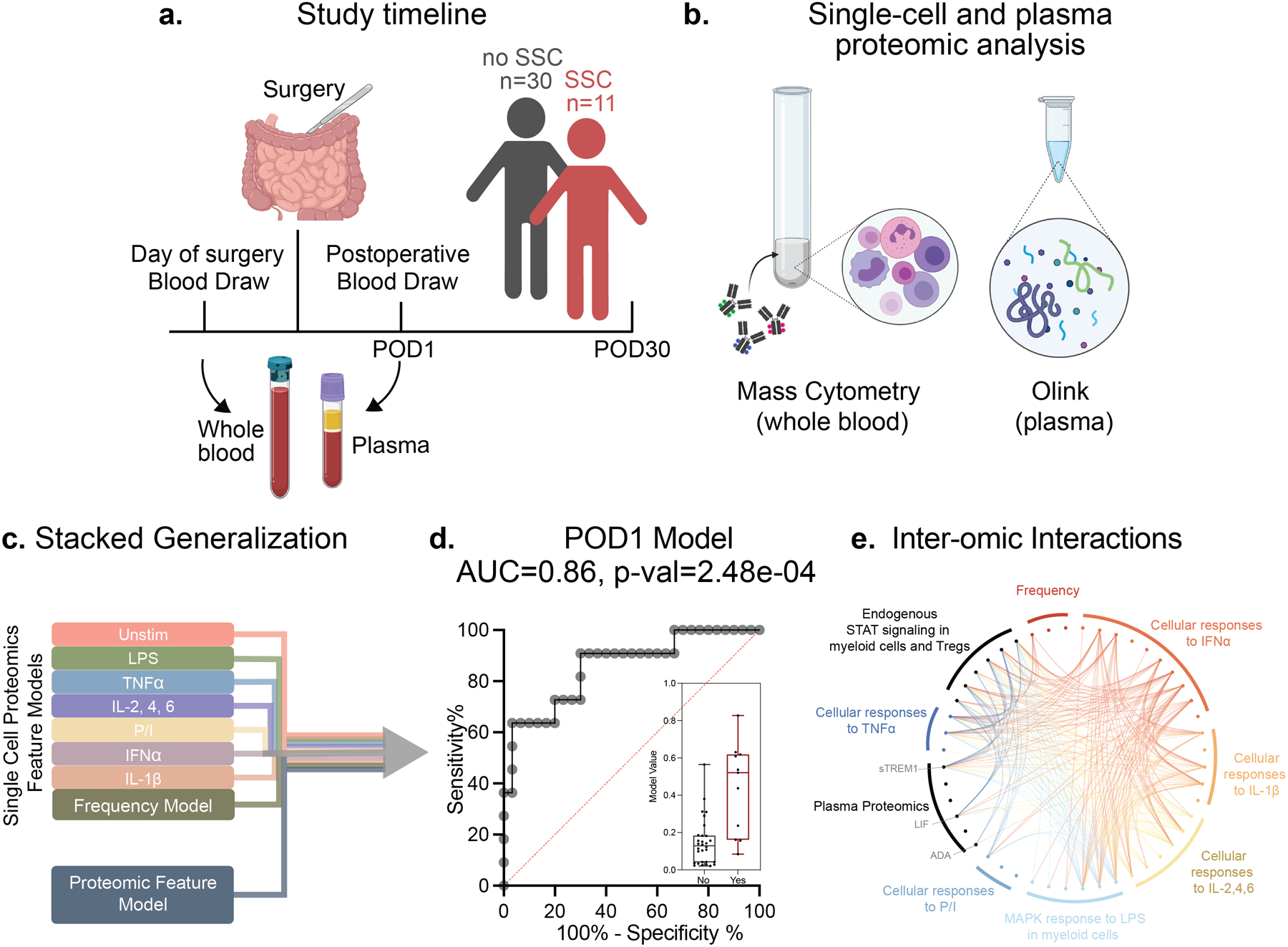
(a) Of the 41 patients enrolled in the study 11 patients developed SSCs within 30 days of surgery while 30 patients did not. (b) Whole blood samples collected on the day of surgery (DOS) prior to induction of anesthesia and on POD1 were stimulated with lipopolysaccharide (LPS), tumor necrosis factor (TNF)α, interleukin (IL)-2,4,6 cocktail, PMA/Ionomycin (P/I), interferon (IFN)α, IL-1β, or left unstimulated (Unstim) for analysis by single-cell mass cytometry. Plasma samples were analyzed using the Olink multiplex proteomic platform. (c) Elastic Net (EN) penalized linear regression models differentiating patients with and without an SSC were trained for individual data layers (plasma proteome, immune cell frequency, single-cell endogenous signaling, and signaling responses to each of six ex vivo stimulations) before integration of all nine data layers using a stacked generalization (SG) method. (d) The SG model output was evaluated using a leave-one-out cross validation (LOOCV) procedure. The receiver operating characteristic (ROC) analysis (area under the curve, AUC = 0.86, p = 2.5e-04), median values, and interquartile range of model outputs (boxplot, p = 2.48e-04, two-sided Mann-Whitney rank-sum test) are shown. (e) Chord diagram depicting correlation between model features of different data layers (Spearman R>0.5). Data layers are highlighted using the color scheme in (a). All model features (fold >2) are listed
