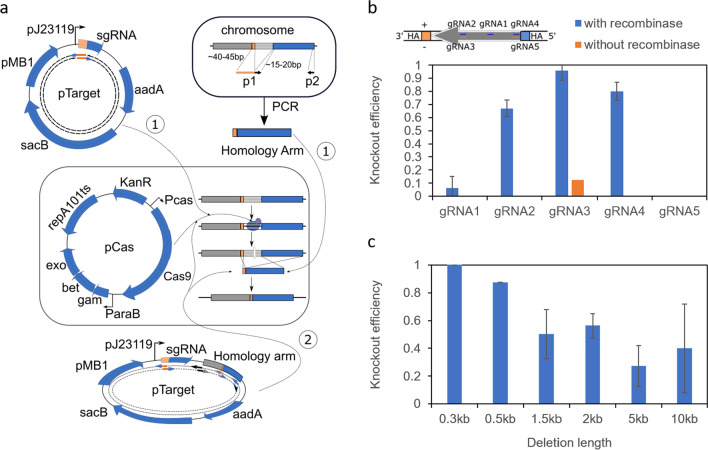
Fig. 1.
The modified CRISPR-Cas9 based genome deletion in the E. coli BL21 strain. a Schematic representation of the modified two-plasmid system
adapted from Jiang et al. [6]. Two different methods have been tested. Both requires the pCas plasmid being transformed into E. coli cell first. Subsequently, the pTarget plasmid with homology arm either as PCR fragments (method 1) or carried on pTarget plasmid (method 2) were transformed into the cell. For method 1, the homology arm is obtained with a simple PCR step where the forward primer 1 (p1) carries the upstream (40–45 bp) homologous sequence fused with the downstream (15-20 bp) homologous sequence for priming, and the reverse primer 2 (p2) is about 15–20 bp targeting 500 bp downstream of p1 priming sequence. The total length of p1 primer is 60 bp to ensure efficient synthesis and PCR. The gRNA sequence is changed with restriction-free cloning method [23]. In total, 4 primers were used for each target gene modification. For method 2, to clone the pTarget plasmid, 4 PCR fragments with 8 primers are used to assemble the plasmid. b Knockout efficiency for CRISPR-Cas9 method 2 in BL21 cell targeting the adhE gene. Five different gRNA designs were tested where their targeting positions are illustrated. Knockout efficiency is calculated based on the number of colonies with successful deletion over the total number of colonies tested. c Knockout efficiency for deleting various lengths from the BL21 genome using gRNA3 targeting adhE region. All the efficiencies were obtained through replicate experiments