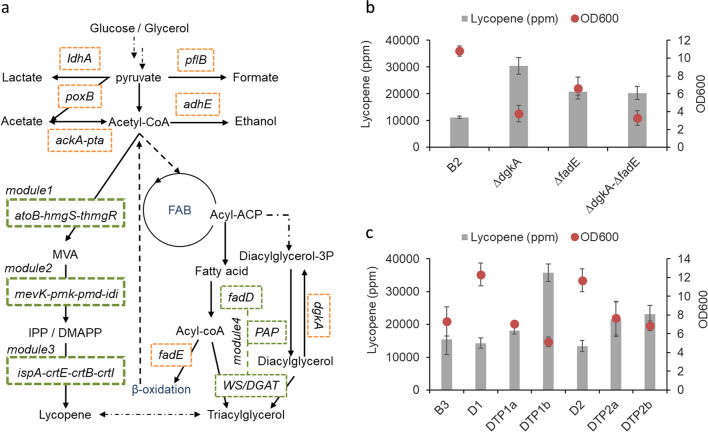
Fig. 5.
Engineering the triacylglycerol (TAG) pathway to increase lycopene production. a). Schematic representation of the TAG pathway and lycopene biosynthetic pathway. The two pathways are grouped into 4 modules, which is boxed with green dash lines (Table 2). The genes boxed in orange dash lines are targets to be deleted. The abbreviations are as follows. adhE alcohol dehydrogenase, ldhA lactate dehydrogenase, poxB pyruvate oxidase, pflB pyruvate-formate lyase, pta phosphate acetyltransferase, ackA acetate kinase, atoB Acetoacetyl-CoA thiolase, hmgS HMG-CoA synthase, thmgR truncated HMG-CoA reductase, mevk mevalonate kinase, pmk phosphomevalonate kinase, pmd mevalonate pyrophosphate decarboxylase, idi IPP isomerase, ispA FPP synthase, crtE GGPP synthase, crtB phytoene synthase, crtI phytoene desaturase, FAB fatty acid biosynthesis; fadD long-chain-fatty-acid—CoA ligase, PAP phosphatidic acid phosphatase, WS/DGAT wax ester synthase/diacylglycerol acyltransferase; dgkA, diacylglycerol kinase; fadE, acyl-coA dehydrogenase. b Specific lycopene yield and biomass of E. coli when dgkA and/or fadE were deleted. c Specific lycopene yield and biomass of E. coli when TAG pathway genes were overexpressed. All the measurements are averaged triplicates with a standard error bar shown in the figure