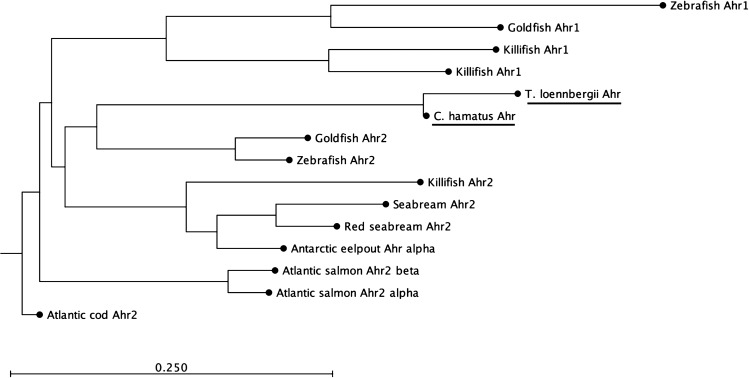
Fig. 2.
Phylogenetic analysis of fish Ahr amino acid sequences. The amino acid sequences were aligned using CLC Main Workbench. The phylogenetic tree was constructed by the neighbour-joining method using CLC Main Workbench, with the branch lengths corresponding to the evolutionary distance between sequence clusters. The GenBank accession of the sequences are as follows: seabream Ahr2 #AAN05089, killifish (Fundulus heteroclitus) Ahr1 #O57452, killifish (Fundulus heteroclitus) Ahr2 #AAC59696, goldfish (Carassius auratus) Ahr1 #ACT79400, goldfish Ahr2 #ACT79401, zebrafish (Danio rerio) Ahr1 #NP571103, zebrafish (Danio rerio) Ahr2 #NP571339, red seabream (Pagrus major) Ahr2 #BAE02825, Atlantic salmon (Salmo salar) Ahr1 #NP_001117158.1 Atlantic salmon (Salmo salar) Ahr2α #NP_001117156, Antarctic eelpout (Pachycara brachycephalum) Ahr2α #KY747528, Atlantic cod (Gadus morhua), Chionodraco (Chionodraco hamatus) Ahr2 and Trematomus (Trematomus loennbergii) Ahr2