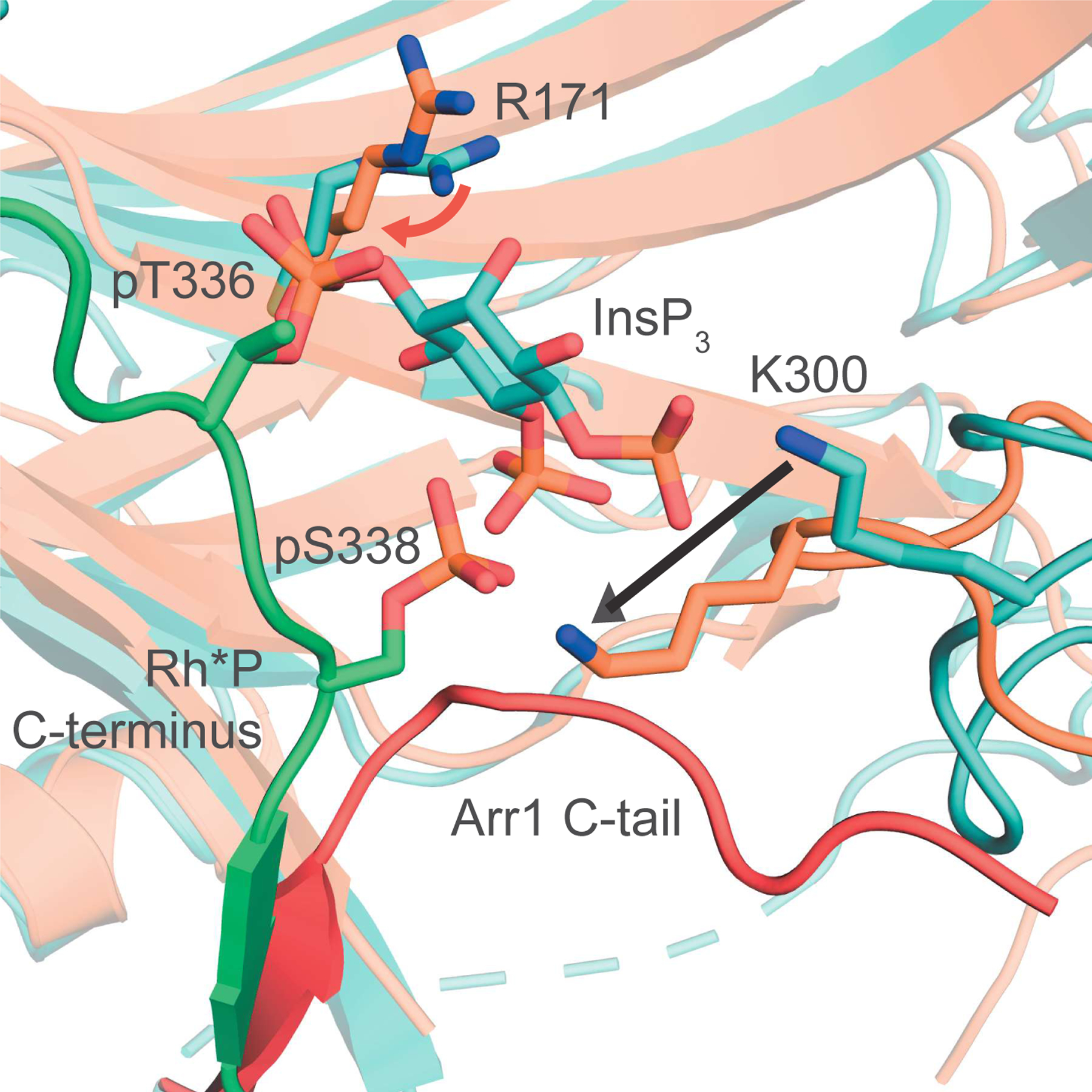
Figure 6. InsPs are present in the vertebrate retina and bind Arr1.
(A) PAGE analysis shows the presence of InsP5 and InsP6 in bovine retina homogenates. Amounts of standards (nmol) are noted at the top. 25% of the sample extracted from 25 bovine retinas was loaded in the “retina” lane. (B-C) Single cell RNA-sequencing (scRNA-seq) heat map depicting absolute (unscaled, B) and relative (scaled as a fraction of the absolute expression value in the retinal cell type with maximal expression for each gene, C) expression values of PP-InsP enzymes by cell type in wild-type (WT) murine retina (Hoang et al., 2020). MC, Müller cell; BC, bipolar cell; AC, amacrine cell; HC, horizontal cell; As, astrocyte; EC, endothelial cell; RPE, retinal pigment epithelium. (D) In situ hybridization analyses of WT murine retinal cryo-sections showing Ppip5k1 and Ip6k2 message throughout the retina with strongest signals in the photoreceptor outer nuclear layer (ONL) (n = 3, representative image shown). The section is co-stained with peanut agglutinin (PNA) and DAPI to demarcate cone photoreceptors and nuclei, respectively. INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar, 50 μm. (E-F) Affinity measurements of natural (E) and synthetic (F) InsPs for Arr1 performed using a fluorescence quenching assay (n=3, mean ± SD). The data in E and F were corrected to account for ligand depletion and the curves based on a one-site binding model.
