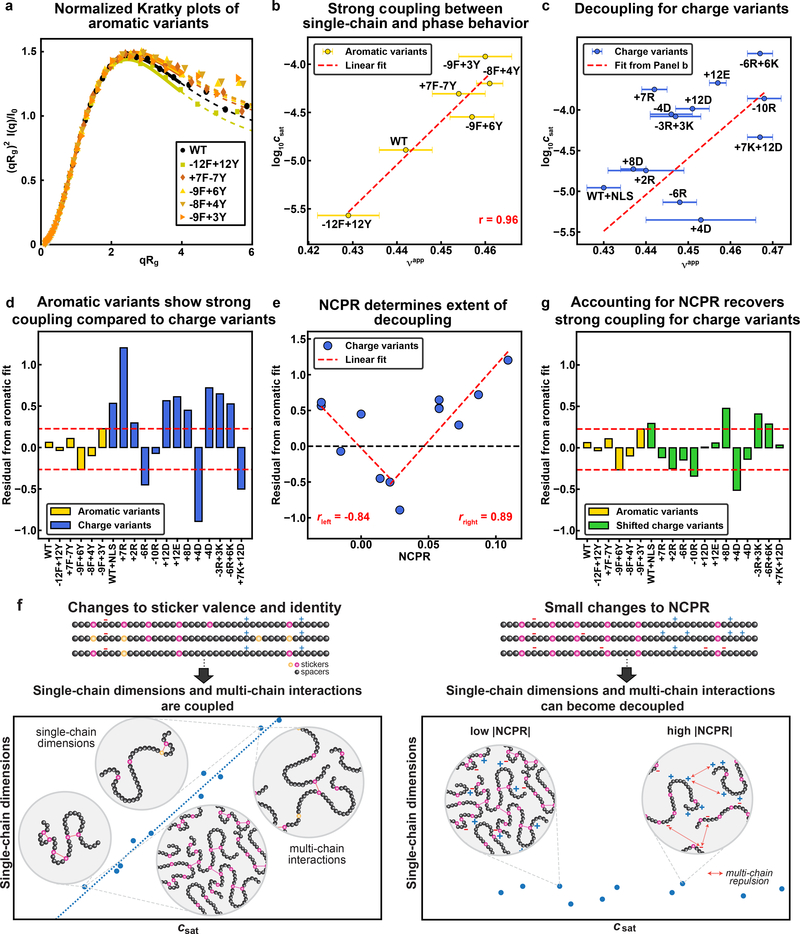
Fig. 6: Mean-field electrostatic effects can disrupt the strong coupling between driving forces for single-chain contraction and phase separation.
(a) Normalized Kratky plot of SEC-SAXS data for aromatic variants of A1-LCD. Solid lines are fits to an empirical molecular form factor (MFF). Dashed lines show the predicted behavior at larger q values, where the experimental data are noisy. (b) Correlation between log10(csat) and inferred values of νapp for the aromatic variants. Red dashed line is the linear fit to the data. (c) Correlation between log10(csat) and inferred νapp values for the charge variants. The dashed line is the linear fit of the aromatic variants from (b). (d) Residuals from the correlation in (b) for all variants. Data for charge variants are shown using unmodified and shifted csat values as in (c) and (e), respectively. Red dashed lines indicate the bounds of the aromatic residuals. (e) Residuals from the correlation in (b) versus NCPR for the charge variants. Red dashed lines show two linear fits, i.e., for the variants whose NCPR is less than or equal to that of the +7K+12D variant and greater than or equal to that of the +7K+12D variant on the left and right, respectively. (f) Residuals from the linear correlation of log10(csat, shift) vs. νapp for charge variants. Here, csat, shift refers to the csat values that are shifted based on the linear fits in (e). Red dashed lines indicate the bounds of the aromatic residuals. (g) Schematic showing strong coupling of single-chain dimensions and driving force for phase separation across variants titrating sticker strength and valence, but the potential for decoupling for variants with small changes to NCPR. Error bars associated with νapp values in (b) and (c) represent uncertainty from fitting SAXS data to MFFs. Saturation concentrations were measured independently at least 3 times for each variant.