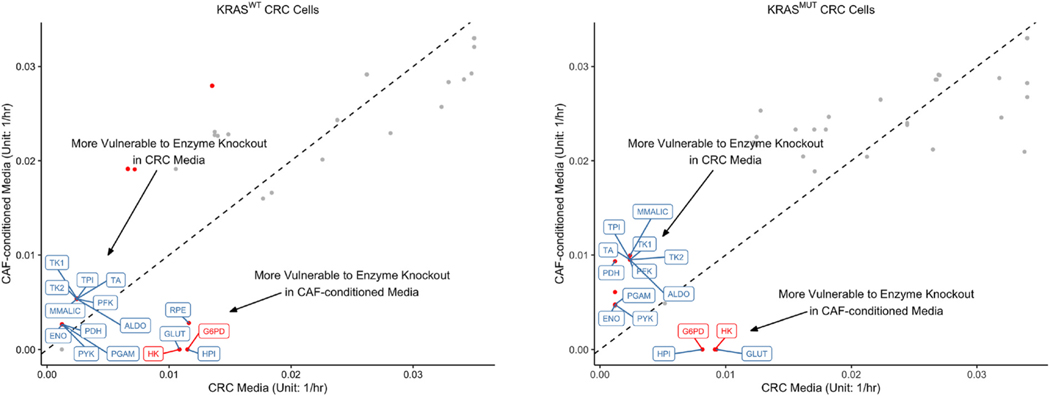
Fig. 9.
Predicted maximal biomass growth rates upon gene deletions. uFBA was used to predict the maximal growth rates for KRASWT and KRASMUT CRC cells upon gene deletions. The x- and y-coordinates of each point represent the median maximal biomass growth rates of CRC cells cultured in CRC media and CAF-conditioned media, respectively when the same enzyme is inhibited. The dashed diagonal line represents equal median rates in both media. Gene deletions resulting in significantly different biomass growth rates between CRC media and CAF-conditioned media are represented by red dots (adjusted p-value < 0.001, fold change > 2). Enzymes belonging to glycolysis, PPP, the TCA cycle, and glutaminolysis are labeled.