Figure 4.
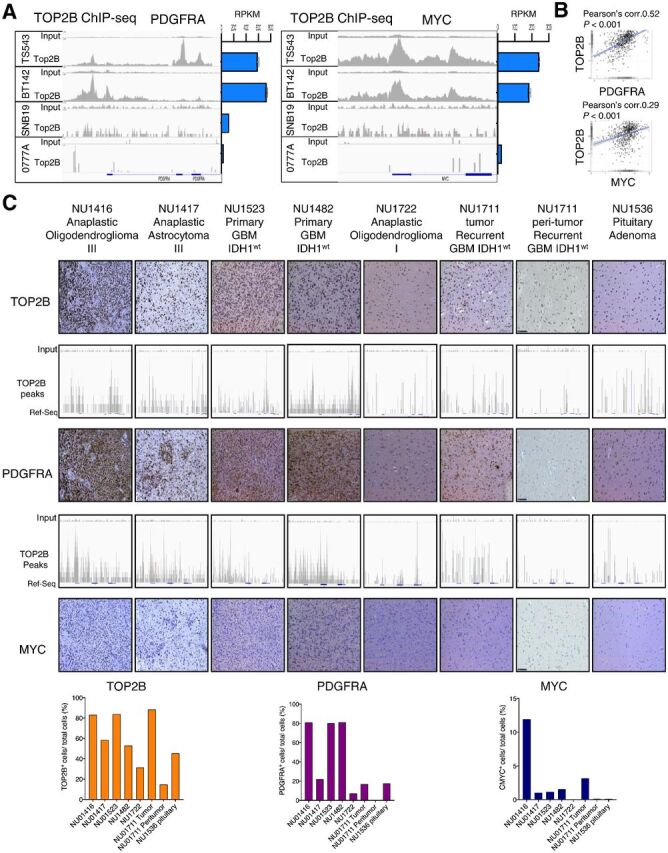
TOP2B expression and genomic localization at PDGFRA and MYC loci is associated with the expression of these genes in human gliomas. A, ChIP-seq for TOP2B native binding (DMSO ChIP; left) and RNA-seq data (right) shows genomic binding and expression of PDGFRA (left) and MYC (right) across human glioma cell lines. ChIP-seq tracks presented are representative of the following experimental replicates: TS543 (n = 2), BT142 (n = 2), 0777A (n = 1), and SNB19 (n = 2). B, Correlation of expression between TOP2B versus PDGFRA mRNA (top) and TOP2B vs. MYC mRNA (bottom) in human gliomas (including low-grade gliomas and GBM) from TCGA GlioVis portal http://gliovis.bioinfo.cnio.es/) determined by Pearson coefficient. C, ChIP-seq for TOP2B at the PDGFRA and MYC loci performed in a set of different glioma human specimens and a sample of peritumoral brain, as well as a pituitary adenoma paired with expression of these proteins determined by IHC (top), and is respective quantification shown in bar plots (bottom). Expression of these proteins were determined by IHC. The slides were quantified for the positive cells and normalized per total cell number for TOP2B, PDGFRA and MYC using HistoQuest version 6.0 software (TissueGnostics). Same y-axis scale was used for input and for the corresponding ChIP track in all cases.
