Abstract
Circadian rhythms are daily cycles in biological function that are ubiquitous in nature. Understood as a means for organisms to anticipate daily environmental changes, circadian rhythms are also important for orchestrating complex biological processes such as immunity. Nowhere is this more evident than in the respiratory system, where circadian rhythms in inflammatory lung disease have been appreciated since ancient times. In this focused review we examine how emerging research on circadian rhythms is being applied to the study of fundamental lung biology and respiratory disease. We begin with a general introduction to circadian rhythms and the molecular circadian clock that underpins them. We then focus on emerging data tying circadian clock function to immunologic activities within the respiratory system. We conclude by considering outstanding questions about biological timing in the lung and how a better command of chronobiology could inform our understanding of complex lung diseases.
Keywords: circadian, rhythm, clock, lung, pulmonary, inflammation
INTRODUCTION
Around the year 100 CE, a Greek physician named Arataeus of Cappadocia recorded the earliest account of an asthma exacerbation that has survived to modern times (1). Detailed in his narrative are the cardinal symptoms of asthma, including chest tightness, cough, and shortness of breath. However, also woven into Arataeus’ description was an observation about timing: Asthma symptoms tended to occur during the night or during sleep (2). What was true about asthma in Arataeus’ time remains true today: Asthmatic patients tend to develop exacerbations during the night, and nocturnal symptoms constitute part of the modern definition of asthma (3–5). We now recognize that many diseases are defined not just by stereotypical symptoms but also by characteristic times of day when those symptoms become apparent, a phenomenon called a circadian rhythm.
Circadian rhythms are daily variations in biological function that have been recognized for centuries to be present in virtually all organisms. The last several years have seen a new enthusiasm for exploring how circadian rhythms affect disease, thanks to the recent discovery of their genetic and molecular underpinnings. Across multiple biomedical disciplines, researchers are revisiting ancient observations about the temporal character of physiology and disease and translating them into molecular terms.
In this focused review, we examine how the new molecular paradigm for circadian rhythm generation is being applied to respiratory disease research. Our specific emphasis is to consider emerging evidence connecting complex immunologic activities such as inflammation to circadian timing within the lung. We conclude by considering what circadian gating of immunity might imply about the fundamental biology of lung disease.
RHYTHMS, CLOCKS, AND HOURGLASSES
Biological rhythms are any predictably repetitive activity observed in living things. They are every-where in nature and occur across diverse time frames: from the millisecond-scale rhythms of action potentials to 17-year rhythms of cicada reproduction (6). However, daily circadian rhythms receive special attention because of the significant impact of the day-night cycle, caused by the rotation of the earth about its axis. The ability to anticipate daily environmental changes represents a powerful selective advantage, and model organisms whose circadian rhythms are genetically disrupted tend to be less reproductively fit than wild-type counterparts (7–9). What exactly about circadian rhythms makes organisms more fit is difficult to define because the phenomenon is so ubiquitous: from behavioral sleep-wake cycles to the rhythmic abundance of individual mRNA species. Adding to the confusion is that many circadian rhythms provide no obvious selective advantage, for example, daily rhythms in the efficiency of antigen presentation (10–12). Some circadian rhythms are not anticipatory at all: For example, rhythms in cortisol seem to function as an “ignore me” signal to prevent unwanted adipogenesis rather than to peg the effect of this hormone to a specific time (13). Finally, some ill effects of interfering with circadian rhythms appear to localize to either the embryo or the uterine environment, a place of relative environmental constancy (14). Regardless of why circadian rhythms evolved, they are clearly imbedded in the operating system of organisms, a framework around which the complex and disparate biological processes that constitute physiology are organized. It is telling that interfering with circadian rhythms through either genetic or environmental manipulation is registered by organisms as stressful and elevates systemic inflammatory markers (15, 16). Over the long term, chronic circadian disruption in humans and model organisms is associated with increased rates of heart disease, metabolic syndrome, and cancer (including lung cancer) (17, 18). Clearly, circadian rhythms are medically relevant, including, as we detail below, in the context of respiratory disease.
In principle, circadian rhythms can be generated via two mechanisms (19). They can be a response to a fixed daily event such as sunrise, but without that daily cue they would fade away (an hourglass mechanism). Alternatively, they can be generated by a self-sustaining internal chronometer that is not reliant on external stimuli for their perpetuation (a clock). Through a series of classic experiments conducted over centuries (20), circadian rhythms were shown to emanate from an internal biological clock with certain universal characteristics. First, circadian clocks have an intrinsic periodicity that is slightly shorter or longer than 24 h, depending on the species. It can be observed by removing organisms from cyclical light and food cycles (this is called free running). Second, the phase of the circadian cycle is programmable by environmental cues such as light and food availability, a phenomenon known as entrainment. Whether exposure to such cues (or zeitgebers) causes a circadian rhythm to spring forward or fall behind depends on where in the cycle the organism is when it encounters the cue (this is called a phase-response curve). Finally, the intrinsic periodicity of circadian rhythms is independent of ambient temperature (temperature compensation) (19). Technically a biological rhythm is circadian only if it satisfies all of the above properties (21). However, some biological rhythms labeled as circadian in the literature have not always satisfied the formal definition. While many of these activities may not be directly driven by the circadian clock, they are often coupled to a process that is. For example, daily variations in the intestinal microbiome of mice depend on circadian feeding patterns but are not themselves directly clock driven (22). More germane to the respiratory system, oxygen consumption is directly clock driven in rodents, but rhythms in CO2 excretion are a product of feeding patterns (23). The key point is that the day-to-day temporal organization of organisms represents a partnership between circadian clock-driven processes and hourglass processes that are coupled to them by specific circadian outputs such as feeding behavior.
CIRCADIAN SWINGS IN LUNG PHYSIOLOGY
Circadian rhythms in respiratory function were long appreciated by physicians, but a physiologic description had to await accurate lung mechanics measurements developed in the twentieth century. Three aspects of respiratory system function were noted by physiologists to have a circadian rhythm. The first is a daily oscillation in airway caliber, reflected in airway resistance measurements such as forced expiratory volume in 1 second (FEV1) or peak expiratory flow rate (24). Airways are at their narrowest (and therefore confer the highest resistance and lowest peak flow) in the early morning between 2–5 am, precisely the interval when asthmatics have the highest likelihood of respiratory symptoms (25). Nighttime increases in airway resistance are further magnified in subjects who are permitted to sleep (26, 27), perhaps due to increased parasympathetic tone. The degree of variation in airway resistance in healthy humans is too small to cause symptoms but becomes clinically impactful when airway diseases such as asthma, chronic obstructive pulmonary disease (COPD), or chronic cigarette smoking are superimposed (28). This magnification of circadian variation can even be seen in children exposed to secondhand smoke who presumably lack chronic airway remodeling (29).
The second classic rhythm in respiratory physiology is the ventilatory response to hypercapnia (elevated CO2 blood levels). Humans are relatively tolerant of CO2 buildup in the early morning, corresponding to the same times of day when airway resistance is highest (30). This arrangement probably minimizes the work of breathing while asleep for healthy individuals but has an important pathological drawback: It predisposes humans to develop sleep apnea (31).
A final classically described circadian rhythm in the human lung is pathological in nature and involves the airway response to an inhaled allergen. Asthmatics challenged with an allergic trigger develop bronchospasm in a biphasic manner: an early asthmatic response that occurs within an hour of encountering antigens and typically spontaneously resolves, followed by a late asthmatic response characterized by increased airway resistance and inflammatory cell infiltration into the airway several hours later (32). The dose of allergen needed to trigger acute bronchospasm varies with the time of day, with allergens being maximally potent if given at night (33–35). However, regardless of the time of trigger exposure, the late asthmatic response appears to be gated to the early morning hours that coincide with the normal circadian nadir in airway caliber (32). The late asthmatic response can recur for several subsequent nights after an asthma trigger (36) and is accompanied by rhythmic increases of lymphocyte infiltration into the airspaces (37). To summarize, the human respiratory system exhibits circadian variations in airway mechanics, ventilatory control, and immune function. The clinical importance of these rhythms has long been appreciated, but until recently we lacked the tools to understand the molecular biology behind this phenomenon. That all changed with the discovery of circadian clock genes.
TO THE BEAT OF A CIRCADIAN DRUMMER
The current explosion in circadian rhythm research arose from mutational screens that identified genes important for circadian patterns of activity in flies and rodents (38). The phylogenetically conserved genes cloned from these screens (clock genes) proved to be bHLH-type transcription factors or interacting partners of these transcription factors that collectively regulate transcriptional activity near E-box promoter motifs (39). The current molecular clock model in mammals (Figure 1) situates a transcription factor heterodimer composed of the proteins BMAL1/ARNTL1/MOP3 and CLOCK at the core of the pathway (39). BMAL1/CLOCK is a histone acetyltransferase and promotes gene expression by making the chromatin landscape around E-box-containing genes more permissive to transcription as opposed to directly recruiting RNA polymerase (40). Among the hundreds of E-box-containing genes transactivated by the core complex are the negative regulatory factors per1–3 and cry1–3 whose gene products bind to and inhibit BMAL1/CLOCK activity. Other clock genes, such as rev-erbα,β and rorα−γ, reciprocally regulate bmal1 and clock gene expression (39). The cumulative result of all this feedback is that the expression of circadian clock genes oscillates with roughly a 24-h periodicity, thus in a sense biochemically encoding the time of day. Several targets of molecular clock regulation are themselves pleiotropic transcription factors, for example, PAR-domain basic leucine zipper transcription factors (DBP, TEF, and HLF) and peroxisome proliferator–activated receptors (PPARs) (41, 42). Through these downstream effectors as well as others, the molecular circadian clock imparts a daily temporal structure on gene expression and, by extension, protein and metabolite abundance. The system contains a high level of genetic redundancy: A deletion of at least two per, cry, or rev-erb genes are needed to inhibit behavioral circadian rhythms in mice. The one exception is bmal1, where deletion of this single gene appears to break the clock (43). Remarkably, mutations that shorten or lengthen the periodicity of per and cry gene expression provoke corresponding changes to the periodicity of organism activity patterns, including humans (44–46). Clock genes are a rare example where the dynamic behavior of a gene scales directly up to the dynamic behavior of an organism.
Figure 1.
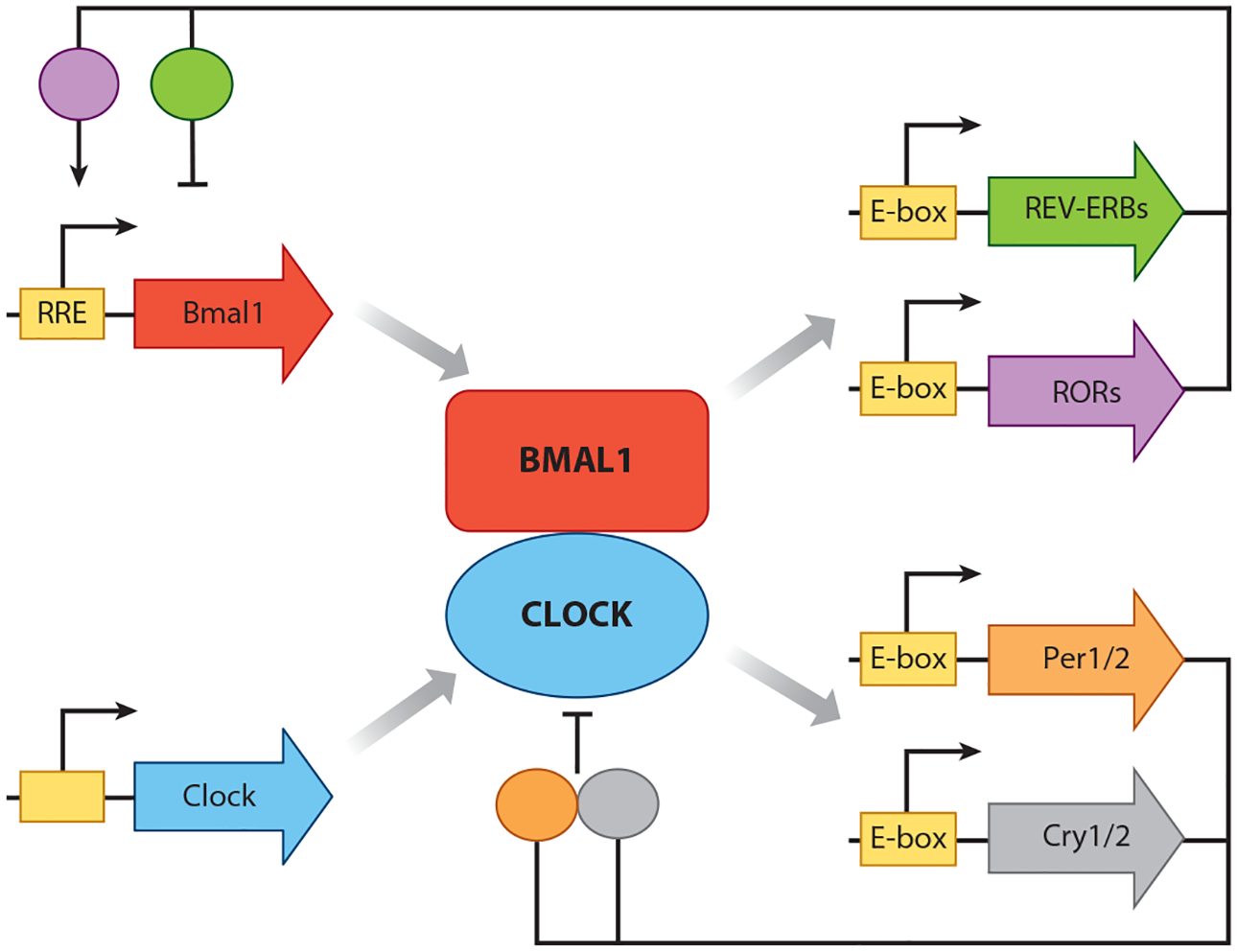
Schematic depiction of the core circadian clock for mammals. Clock genes are depicted as arrows, and their protein products are represented as ovals or circles. Yellow boxes depict E-boxes or ROR-responsive elements (RREs) that are bound by BMAL1/CLOCK or REV-ERB/ROR proteins, respectively. For simplicity, we have not depicted accessory proteins such as casein kinase 1δ/ε (CK1δ/ε) that serve to adjust periodicity and phase of the clock but are not central to rhythm generation (39). Note that there are additional molecular clock constituents (also not depicted) that are expressed selectively in the central nervous system, for example, NPAS2 (a functional homologue of CLOCK) and DEC1/2 (that provide additional negative feedback to BMAL1/CLOCK) (68, 142).
As powerful as the molecular clock pathway is as a model, it is worth pointing out some observations that are hard to reconcile with the canonical paradigm. The oscillation of some clock genes such as bmal1 are dispensable for maintaining circadian rhythms in activity (47). Moreover, not all clock-driven circadian rhythms in nature require transcription: Cyanobacterial circadian rhythms are driven by cyclical phosphorylation of a divergent group of proteins (48), while rhythms in reactive oxygen species (ROS) generation in erythrocytes occur in the absence of a nucleus (49). Nor are clock genes necessarily the sole seat of temporal information within nucleated cells: Mitochondrial respiration exhibits circadian variation, and depleting cells of mitochondrial DNA damps clock gene oscillations (50). Finally, circadian gene expression in cells grown in vitro is much less robust than in vivo, suggesting that cell intrinsic clocks are not enough by themselves to recapitulate the full temporal dynamics of organisms (51, 52). It is probably too simplistic to say that clock genes are in charge of circadian rhythms in cellular physiology; rather, they represent a genetic cog in a complex oscillatory system, including all classes of biomolecules within cells as well as communication between cells (53, 54). Nevertheless, clock genes and the paradigm built around them represent a major advance that allows us to use modern tools of genetics to examine the circadian contributions to lung disease.
In principle, any feedback loop, transcriptional or otherwise, can produce oscillations. It is reasonable to ask why clock genes, as opposed to any other transcription factor network, became the drivers of circadian rhythms in higher organisms. One potential reason is that the clock network has extensive cross-talk with multiple aspects of cellular physiology. For example, clock gene products interact extensively with nuclear receptors, 50% of which exhibit circadian variations in expression (in fact, the REV-ERB and ROR families of clock proteins are themselves classified as nuclear receptors) (55, 56). The natural ligands of the nuclear receptor family are diverse small metabolites such as heme, sterols, and retinoids (57, 58). As a result, clock gene products are functionally receptive to the metabolic status of cells. Another aspect of clock proteins is their promiscuous protein–protein interactions that connect them to diverse regulatory systems. For example, BMAL1 is not just a nuclear transcription factor but also interacts with the translation machinery in the cytoplasm (59). PER2 interacts with the mTOR complex, giving it a connection to nutrient sensing and protein catabolism (60). CRY1 regulates gluconeogenesis in part through interactions with G protein–coupled receptors (61). Clock gene products also appear to bind to other nonclock transcription factors, both ubiquitously expressed proteins such as nuclear receptors and tissue-specific factors such as HNF6 (62, 63). Finally, clock gene products are sensitive to numerous forms of covalent modifications that enable diverse feedback regulation, including phosphorylation, SUMOylation, acetylation, and ubiquitination (64). Taken together, the clock gene network combines a deep embedding into cellular metabolism with programmability, making it ideal for aligning cellular physiology with the solar day.
ONE CLOCK TO RULE THEM ALL
Because the most easily observable circadian outputs in mammals are behavioral, one might think that the molecular clock would operate selectively in the central nervous system. However, it became clear early on that clock genes are expressed in most nucleated cells in mammals, allowing individual peripheral tissues to keep time, including in the lung (65). Under standard tissue culture conditions, the phases of individual cellular clocks are randomly out of phase with each other (66, 67). For the molecular clock to be useful to organisms composed of billions of cells, cell-autonomous clocks must somehow be aligned. In mammals, cellular clocks in the periphery are kept in register by a small region of the ventral hypothalamus containing about 20,000 neurons called the suprachiasmatic nucleus (SCN). The SCN receives afferent innervation about environmental light levels from retinal ganglion cells through the retinohypothalamic tract, which synchronizes the molecular clocks of the SCN cells via cAMP response element binding (CREB) protein (68). These neurons in turn impart rhythmic behavior to other regions of the hypothalamus that regulate pituitary hormone secretion, temperature, autonomic tone, appetite, and arousal (68). The resultant cues working in combination are thought to synchronize the individual cellular clocks of peripheral organs (68). Lesioning the SCN renders mammals behaviorally arrhythmic when they are deprived of light cues, while surgical implantation of an SCN from another organism is enough to restore these rhythms (69). Based on data from mouse liver and lung, 10–15% of normally cycling transcripts can be directly driven by the SCN in the absence of a functioning local clock, including the clock gene per2 (70–72). For all of these reasons, the SCN is often referred to as the master circadian pacemaker.
WHAT DO LUNGS USE TEMPORAL INFORMATION FOR?
One way to infer the role played by the molecular circadian clock in a given organ is to compare its circadian transcriptome to that of other tissues. In the last five years, organ-specific atlases of circadian gene expression that include the lung have been compiled for rodents, primates, and postmortem human specimens (73–77). Overall, 5–20% of transcripts exhibit daily oscillation in expression in any given organ, with 40–80% of all genes exhibiting daily rhythmic expression in some mammalian tissue, depending on the species (73–77). Gauged in terms of the number of oscillating transcripts, the lung ranks as the third most rhythmic solid organ in mice, slightly behind liver and kidney but significantly more rhythmic than the SCN, the master circadian pacemaker (75). This kind of ranking probably understates the circadian character of tissues with high cellular diversity such as the as the lung because cell type–specific signals are likely to be obscured. Although clock genes exhibit conserved rhythmic expression in all organs, this tells us shockingly little about what other genes will oscillate in any given tissue. For example, the lung circadian transcriptome of healthy mice has only a 5–26% overlap with other mouse organs (75). The reasons for why circadian transcriptomes are so context specific are incompletely understood but probably related to cell type–specific differences in chromatin accessibility such that different E-box motifs are available to interact with BMAL1/CLOCK in different tissues (78). Additionally, circadian clock proteins such as REV-ERBα can interact out of network with cell-specific transcription factors, for example, HNF6 in mouse liver, thereby conferring rhythmic activity on tissue-specific gene expression (79). The key point is that the molecular clock does not dictate to cells a specific temporal program in gene expression. Instead, cells harness the temporal information broadcast by the circadian clock to temporally organize processes that are important to their specialized function.
What then does the lung circadian transcriptome tell us about what clocks do in the respiratory system? Intriguingly, functional enrichment analyses suggested a role for the lung peripheral clock in innate and acquired immunity in mice, both in the healthy state and even in the setting of systemic inflammation caused by endotoxemia (Figure 2) (80). This conclusion was bolstered by informatic analyses of a human lung circadian transcriptome that was constructed from biopsy samples using CYCLOPS, a novel machine-learning algorithm (73, 74). Among solid organs in mice, the informatic connection between circadian gene regulation and immunity is arguably most apparent in the lung. Moreover, several members of the lung circadian transcriptome have straightforward clinical correlates: For example, IL18R1, RORα, and Clock are rhythmic in mouse lung and were previously identified as asthma susceptibility loci in various clinical cohorts (81–83). To summarize, the lung circadian clock appears to be specialized for imparting temporal information to the immune response. In order to understand how circadian regulation relates to immunity, we next explore the molecular anatomy of the lung peripheral clock and its properties.
Figure 2.
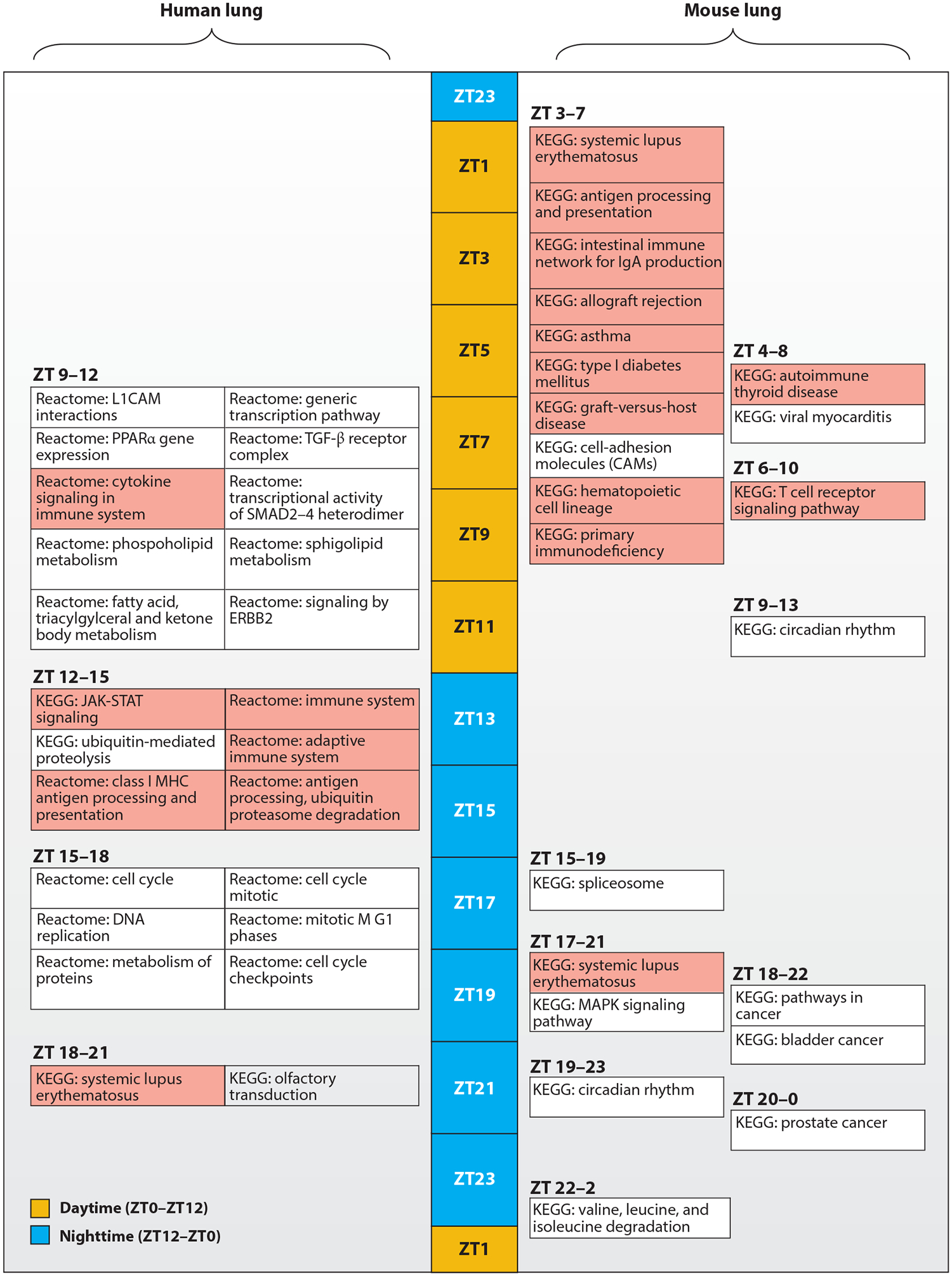
Lung circadian gene expression is specialized for immune regulation. Depicted is a functional enrichment analysis of the human and mouse lung circadian transcriptome synthesized from recently published data (74, 80). Enriched functional enrichment terms are positioned along a 24-h time line expressed in units of zeitgeber time (ZT), where ZT0 represents the onset of daytime (yellow) and Z12 the onset of nighttime (blue). Functional enrichment terms relating to immune function are colored red. In both mice and humans, immune pathways are predominantly enriched at the gene expression level during their respective natural rest phases.
WHAT DO WE KNOW ABOUT THE LUNG PERIPHERAL CLOCK?
Anatomically and physiologically, the lung is composed of two basic compartments: conducting airways that condition the air and transport it to the organ interior and the alveolar space where gas exchange occurs. Both compartments are by themselves complex composite structures, composed of multiple cell types (84). Adding to the diversity, the lung is also a major immune organ, with 15–30% of nucleated cells being leukocytes (85, 86).
At the whole-organ level, clock gene expression is rhythmic in the lung and can be entrained by universal cues such as food, glucocorticoids, and temperature (72, 87, 88). Autonomic tone appears to be an additional synchronizing cue: In rats, vagotomy inhibits circadian rhythms in mucus gland secretion (89). Although cortisol is a strong synchronizer in the lung, it appears to be dispensable for maintaining oscillations in lung clock gene expression, consistent with the clock’s cell-autonomous nature (90, 91). Like most peripheral organs, the lung clock is classified as a weak oscillator in that it can be made to cycle at a relatively wide range of periodicities (20–28 h) (92). Unlike in most organs, clock gene function in the lung is especially sensitive to aging. In one study, lung explants taken from aged mice showed no spontaneous rhythms in per1 expression in 50% of specimens, whereas other solid organs and the SCN were unaffected by old age (93). Lung circadian gene expression is accompanied by a lung circadian metabolome that includes polyamines, nicotinamide, and purine nucleotide metabolites (80). Interestingly, the rhythm characteristics of the lung circadian transcriptome and metabolome are tidally locked: Endotoxin exposure almost identically alters the periodicity and phase distribution of the circadian transcriptome and metabolome in the mouse lung (80).
Based on traditional morphometry and contemporary single-cell RNA sequencing analysis, at least 40 distinct cell types maintain lung architecture in both rodents and humans (84, 94, 95). As such, there is enormous potential for cell-specific control of circadian regulation in the lung. For example, the amplitude of clock gene expression is mechanosensitive but has opposite effects in different lung cell types: Rigid extracellular environments damp clock gene rhythms in lung epithelial cells but enhance their amplitude in fibroblasts (96). Immunofluorescence studies suggested that clock protein expression was concentrated in airway epithelial cells in both humans and rodents, with more spotty expression in alveolar spaces (97). Chemical ablation of club cells, which is the predominant epithelial cell type in the mouse airway, was enough to eliminate circadian rhythms in lung slices as judged by a common reporter system where the coding sequence for luciferase is inserted into the per2 locus (97). Using the same assay, similar effects were produced by deleting bmal1 specifically in airway epithelial cells (90). Together, these data were initially interpreted to mean that airway epithelial cells serve as a kind of local circadian pacemaker within the lung (97). This conclusion has been recently challenged by single-cell RNA sequencing data suggesting that clock genes are expressed in diverse cell types in the lung (94, 95, 98). Moreover, deletion of bmal1 in club cells does not abolish rhythmic clock gene expression in whole lung extracts (72). Additionally, genetic deletion of the glucocorticoid receptor or double deletion of rev-erbα and rev-erbβ in club cells had little effect on rhythms in the alveolar space as judged by the per2-luciferase reporter assay (91, 99). More research is needed to settle whether the airway epithelium provides an instructive role for lung circadian regulation as a whole. Regardless, it is probably safe to assume that different lung parenchymal cell types will have distinctive circadian transcriptomes that fit their specialized roles in respiratory physiology. At present, we have detailed information about clock gene function in only one resident cell type in the lung: club cells, which represent the predominant epithelial cell lining conducting airways in the mouse. In the next section, we review emerging data suggesting a role for the airway epithelial clock in innate immune responses.
THE AIRWAY EPITHELIAL CLOCK: HERALD OF INFLAMMATION
A relationship between circadian rhythms and immune function was first observed decades ago by Halberg et al. (100), who noted that the degree of inflammation elicited by endotoxin depended on when it was administered. Gibbs et al. (90) used inhaled endotoxin treatment to explore the circadian influences in lung inflammation in mice. They found that airway inflammation in response to endotoxin varied with the time of administration, as measured by neutrophil influx and cytokine levels recovered in bronchioalveolar lavage (BAL) fluid. These investigators had previously shown that deleting bmal1 or rev-erbα resulted in excess cytokine secretion by macrophages upon stimulation (101). However, conditional deletion of bmal1 in myeloid cells did not alter the circadian response to inhaled endotoxin, contrary to expectation. Instead, temporal variation in endotoxin responses were sensitive to bmal1 deletion in airway epithelial (club) cells (90). Airway epithelial bmal1 deletion also produced a magnified inflammatory reaction to inhaled endotoxin attributed to dysregulation of CXCL5 expression, a neutrophil chemoattractant. They showed that bmal1 serves to target the glucocorticoid receptor to the cxcl5 promoter, thereby inhibiting transcription of this gene during the night (90). These results were the first to pinpoint a role for airway epithelial cells in temporally organizing acute airway inflammation. Recent work by Zhang et al. (72) suggested that circadian clock function in airway epithelial cells is also needed to mitigate chronic inflammation even under basal conditions: Middle-aged mice bearing club cell deletion of bmal1 spontaneously develop airway-centric fibrosis (72). To help explain why, the authors generated circadian transcriptomes from microdissected airway epithelium obtained from wild-type and club cell-conditional bmal1 knockout mice. Surprisingly, bmal1 deletion did not lead to a loss of circadian gene expression in club cells, but rather a reprogramming of their circadian transcriptome such that immune pathways such as antigen presentation became more enriched at the expense of metabolic pathways (72). Interestingly, a similar proinflammatory reprogramming of circadian gene expression was observed in whole mouse lungs after intraperitoneal injection of endotoxin (80). More recently, circadian clock proteins such as Clock and REV-ERBα were shown to be rapidly depleted after endotoxin treatment, potentially due to proteasomal degradation (99, 102). It is tempting to speculate that airway epithelial cells are preprogrammed to initiate innate immune activities, and this proinflammatory bias must be actively counteracted by normal circadian clock function. This might help explain why diverse stimuli that disrupt or misalign circadian rhythms in otherwise healthy organisms are quick to increase inflammatory markers (15, 16).
While circadian clock function in airway epithelial cells is important for regulating lung immunity, it is not the whole story. Studies in rev-erbα knockout mice showed that deletion in this downstream effector of bmal1 also led to a loss of temporally gated neutrophil influx after inhaled endotoxin (99). In contrast to bmal1 deletion, whose temporal gating activity localized to airway epithelial cells in this model, the circadian actions of rev-erbα localized to myeloid cells, which have a functional clock of their own (101). At first glance this is a confusing result, but it probably reflects the holistic role biological timing plays in immunity. Inflammation is a complex process involving a large and diverse cast of cells that must come together in the right places and at the right times to protect the host. It is logical that circadian clocks in multiple cell types must cooperate to bring about an organized immune response to infection or sterile injury. In the case of the myeloid cell autonomous clock, emerging research suggests a role for it in regulating leukocyte trafficking, the subject of the next section.
CIRCADIAN LEUKOCYTE TRAFFICKING IN THE LUNG
Initial analyses of circadian gene expression in mouse lung noted that prototypical leukocyte lineage genes (such as cd19 for B cells) are rhythmically expressed, reflecting oscillations in lung leukocyte number (80). Importantly, rhythmic leukocyte trafficking in the lung is not unique to the healthy state but occurs during systemic inflammation as well (80). It is worth mentioning that rhythms in leukocyte trafficking are an organism-wide phenomenon in both mice and humans (103–105). They represent the sum of several independent processes, all of which appear to be oscillatory in isolation (Figure 3), including the rhythmic release of cells from the bone marrow (106), rhythms in leukocyte homing to organs (104, 107), consumption by resident phagocytic cells (108), and egress via tissue lymphatics to area lymph nodes (109). A common theme is that the timing of leukocyte trafficking is a joint venture between circadian clocks in the leukocyte and those in endothelial cells of peripheral organs, which together align the expression of receptor–ligand pairs critical for trafficking steps (104, 107). Leukocyte abundance in the lung and other peripheral organs peaks during the daytime in mice, representing the rest phase for this nocturnal animal. However, the key molecules involved in leukocyte trafficking rhythms appear to vary depending on the leukocyte subtype and the destination organ.
Figure 3.
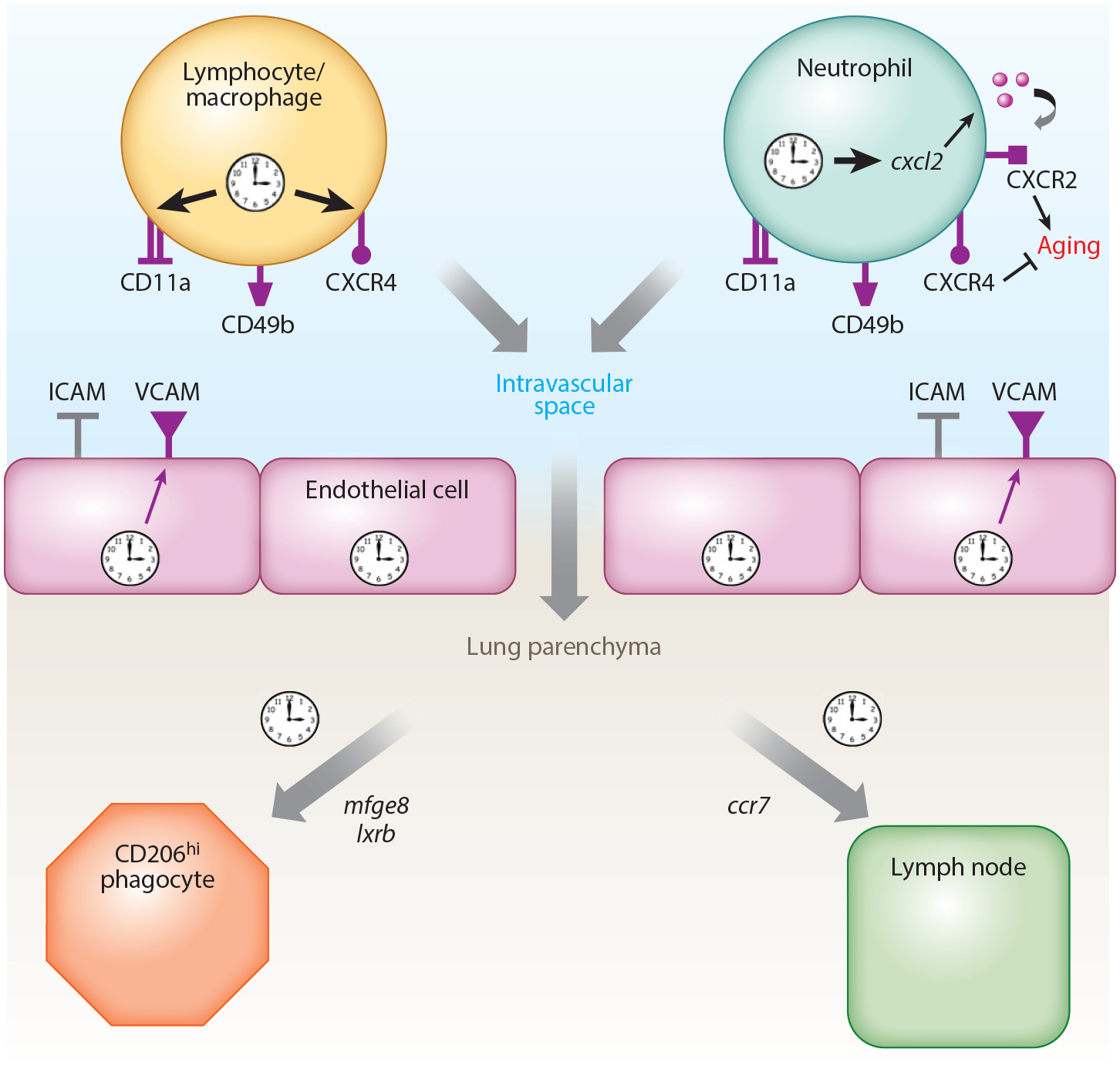
Schematic depicting the circadian control of homeostatic leukocyte trafficking in the lung. The diagram is synthesized from data in several recent publications detailed in this review (107–110). Cell-surface markers important for rhythmic homing of leukocytes are depicted. Receptors whose cell surface expression oscillates in a circadian manner are colored purple. Also depicted are genes important for rhythms in leukocyte removal by interstitial phagocytic cells and for rhythmic homing to area lymph nodes.
Several recent publications have clarified the factors important for the lung homing. For most leukocyte subsets, rhythmic homing to lung is dependent on the endothelial cell adhesion molecules ICAM-1 and VCAM-1, the latter of which is expressed in a circadian manner (107). On the leukocyte surface, CD49b, CD11a, and CXCR4 are important for homing to lung, all of which are expressed rhythmically (107). Cxcr4 is a direct transcriptional target of BMAL1/CLOCK, whose engagement on the promoter of this gene requires ROS production (105). One exception to the above mechanism is neutrophils, whose rhythmic homing is in sync with other leukocyte cell types but employs a different mechanism based on aging. Neutrophils have a short life span (about 1 day), and as they age they exhibit progressive cytoskeletal changes mediated by the loss of CXCR2 signaling as well as depletion of cell-surface CD62L protein (110). Importantly, it is the aged CD62Llow neutrophils that inhabit peripheral tissues during the nighttime in mice under healthy conditions, including the lung (110). Deletion of bmal1 in neutrophils inhibits this aging process because circadian clock function is needed to rhythmically transactivate cxcl2, which contains an E-box and codes for the ligand of CXCR2 (110). Thus, in neutrophils, circadian homing to lung and other peripheral tissues is driven by clock regulation of cytoskeletal alterations and aging rather than through specific cell-adhesion molecules. Surprisingly, neutrophils may have an additional, potentially nonimmune role in regulating lung circadian gene expression. In a recent study, depleting mice of their neutrophils produced a 25% reduction in the number of oscillatory transcripts detected in the lung (111). Thus, immune trafficking appears to be both an outcome of circadian regulation and potentially a feedback regulator that informs temporal programs in the lung.
Once leukocytes arrive in peripheral organs they are cleared in a diurnal fashion by either macrophage phagocytosis (108, 112) or migration to regional lymph nodes (109). It is unclear from published data whether rhythms in lung phagocytosis are intrinsically clock driven or governed by the availability of leukocytes at any given time of day. In the lung, leukocyte clearance appears to be performed by interstitial macrophages rather than alveolar macrophages. Factors regulating phagocytic activity are tissue specific and, in the lung, involve the opsonin Mfeg8 and the nuclear receptors LXRα/β (108). Lymphocyte trafficking from lung to lymph nodes has not been specifically studied but is clock driven in other contexts and was shown to be mediated by lymphocyte rhythms in CCR7 expression (109).
To summarize, data accumulated to date affirm an important role for circadian rhythms in optimizing innate immune responses in the lung. The effects of clocks are evident at multiple levels of the immune surveillance: from rhythms in leukocyte trafficking that provide surveillance for pathogens, to inflammatory cytokine responses at epithelial sites of injury, and to immune cell recruitment and clearance. It is no wonder that circadian rhythms appear to play a role in virtually every lung disease where clock gene function has been examined.
CLOCKS AND LUNGS DISEASE
Asthma
Asthma is a heterogenous inflammatory disease of the airway that remains the most common chronic disease of children in many Western countries (113). Increasingly, the disease is seen as a collection of mechanistically distinct entities that converge on a common phenotype of airway inflammation, remodeling, and episodic wheezing (113). Two clinical aspects tie diverse groups of asthmatics together: the tendency toward nocturnal symptoms and the importance of either infectious or allergic triggers in precipitating attacks. As such, recent efforts have focused on the role circadian clocks play in the response to asthma triggers. Using a mouse parainfluenza virus model, our group found that the amount of acute inflammation after viral challenge depends on the time of inoculation (114). Disruption of circadian function by bmal1 deletion or chronic jet lag led to exaggerated inflammation during acute viral illness. Deletion of bmal1 further induced elevated parainfluenza virus replication in mice and in tissue culture cells (114, 115). The result was significantly worse asthmatic airway remodeling in bmal1-null animals (114). Collectively, these results suggest a role for circadian rhythms in those aspects of asthma that relate to viral triggers. In contrast, clock gene deletion appears to have much less of an impact on allergen-induced lung disease in mouse models. Myeloid deletion of bmal1 led to increased airway eosinophilia after allergen challenge with ovalbumin (116) but in our hands did not exacerbate mucus metaplasia or other overt signs of structural remodeling (J. Haspel, unpublished observations). Similarly, ovalbumin challenge of rev-erbα knockout mice did not yield worse lung remodeling compared to wild-type littermates (99). The relative lack of phenotype in models of atopic asthma is surprising because allergic symptoms in humans have a strong circadian character (117, 118). Common mouse models may be suboptimal for studying the circadian effects of allergic asthma because of species differences in adaptive immunity, the choice of standardized allergens, or the genetic background of the animals (119). Nevertheless, circadian effects are likely to be of medical importance for both allergic and nonallergic asthma. For example, administering inhaled corticosteroids at night appears to achieve asthma symptom control with lower doses of drug, and enforcement of sleep hygiene may improve nocturnal airway resistance in asthmatic adolescents (120, 121). Further examination of how circadian clocks impact asthma phenotypes at a molecular level is warranted.
Smoking-Related Airway Disease
COPD is a chronic inflammatory airway disease usually associated with tobacco smoking that shares overlapping clinical features with asthma, including a tendency to flare at night (122). Like asthma, both COPD and passive secondhand smoke exposure are associated with an increased circadian variation in airway resistance, suggesting a role for circadian clocks in smoking-related lung disease (28, 29). In mice, chronic smoke exposure damps rhythmic bmal1 and clock gene expression and shortens the free-running periodicity of daily activity (123). One mouse-based study examined how chronic smoke exposure affected the clinical severity of influenza A virus (IAV) infection, a common seasonal trigger for COPD (124). They found that smoke exposure worsened acute IAV respiratory infection and, importantly, this phenotype could be recapitulated in the absence of smoking by deleting bmal1 (124). The authors interpreted these data to mean that cigarette smoke produces a hyperactive inflammatory phenotype in airway epithelial cells through suppression of clock gene function (124). Supporting this, Pariollaud et al. (99) showed that rev-erbα knockout mice exhibited moderate mortality after a normally sublethal smoke exposure, and this was associated with elevated lung CXCL1 levels. Taken together, mouse models imply that circadian clock disruption may be an important cause of inflammation in tobacco users and may contribute to respiratory decline in COPD patients.
Fibrotic Lung Disease
Pulmonary fibrosis represents a constellation of progressive lung diseases of unknown etiology in which alveoli are replaced by collagen deposits and knots of proliferating fibroblasts (125). It is controversial whether pulmonary fibrosis is an inflammatory disorder, as immunosuppressants do not arrest disease progression in most patients (125). Recent theories suggest that pulmonary fibrosis begins with a loss or dysfunction of type II pneumocytes (126), perhaps as a function of aging (127). However, the most common murine model for lung fibrosis, bleomycin instillation, does have a strong inflammatory component (128) and implicates circadian clocks in this kind of pathology (129). Even under basal conditions, deletion or mutation of clock genes in mice leads to spontaneous airway-centric fibrosis by middle age, which in the case of bmal1 deletion can be traced to airway epithelial cells (72, 129). Treatment with bleomycin leads to excessive fibrosis in circadian disrupted clockΔ19 mice, an effect attributed to diminished expression of nrf2, a master regulator of antioxidant enzymes (129). Once a lung becomes fibrotic, we know little about what happens to its peripheral circadian clocks. However, overexpression of the profibrotic cytokine TGF-β in cell culture damps bmal1 expression rhythms (130), and extracellular matrix rigidity appears to lower the amplitude of clock gene expression in airway epithelial cells while enhancing rhythms in fibroblasts (96). Given this, fibrotic lungs are likely to have an altered temporal landscape that may have implications for immune functions, such as leukocyte trafficking, in these remodeled organs. Further research is needed to investigate this possibility.
Pneumonia
Given the role that lung peripheral clocks play in limiting airway inflammation, one might think that clock disruption ought to mitigate severe respiratory infections such as pneumonia. However, data from mouse models of bacterial and viral pneumonia suggest the opposite. In mice, pneumococcal pneumonia severity varied with time of infection as measured by lung bacterial load and lung neutrophilia (90). Deletion of bmal1 from airway epithelial cells significantly increased lung neutrophils in infected animals, but this did not result in better bacterial clearance (90). Why increased neutrophilia did not translate into better bacterial control is unclear. In principle, it may mean that airway clock deficiency impedes the initial recognition of bacteria by lung innate defenses, the local milieu produced by defective airway clocks negatively affects neutrophil function, or simply that neutrophil number is already saturating in mouse models of pneumococcal pneumonia and additional neutrophil recruitment is redundant.
Circadian rhythms in viral respiratory illness have so far been examined in mice for parainfluenza virus, which produces bronchiolitis, and for IAV, an important cause of pneumonia in humans. With either virus, acute inflammatory responses, but not the peak viral load, vary with the time of inoculation in wild-type mice (114, 131). Similarly, bmal1 deletion worsens acute lung injury and lung inflammation in response to parainfluenza or IAV (114, 124, 131), which in the case of IAV may be mediated specifically by a disrupted airway epithelial clock (72). Where parainfluenza and IAV diverge is in the effect that bmal1 deletion has on viral replication. Deletion of this key clock gene raises parainfluenza viral titer by 0.5–1 log during acute infection (114), whereas investigators differ on whether bmal1 regulates IAV viral load (131, 132). Moreover, the duration of IAV illness in bmal1 knockout mice appears to be significantly prolonged (72, 114, 124, 131), suggesting that circadian clocks may play a role (immunologic or otherwise) in the resolution of viral pneumonia.
A feared complication in pneumonia survivors is the development of chronic lung disease, a major source of disability and medical costs in patients. Relevant to this, certain mouse strains are prone to chronic, inflammatory lung pathology after parainfluenza or IAV infection, although the histologic picture produced by each virus is different (133, 134). With either virus, deleting bmal1 seems to enhance the amount of chronically remodeled lung tissue postinfection (Figure 4). At present, relatively little is known about how the immune system contributes to the restoration of lung architecture after pathogen clearance has been achieved, but the fact that circadian regulation may be involved represents an important consideration for future study. To summarize, there is strong evidence from animal models that circadian regulation within the lung is important for the immunologic control of acute lung infection and the likelihood of developing chronic lung disease in its aftermath. As with pulmonary fibrosis, any postinfectious remodeling of lung architecture is likely to impose a novel circadian landscape that departs from what has been observed in healthy mice and represents an important area for future study.
Figure 4.
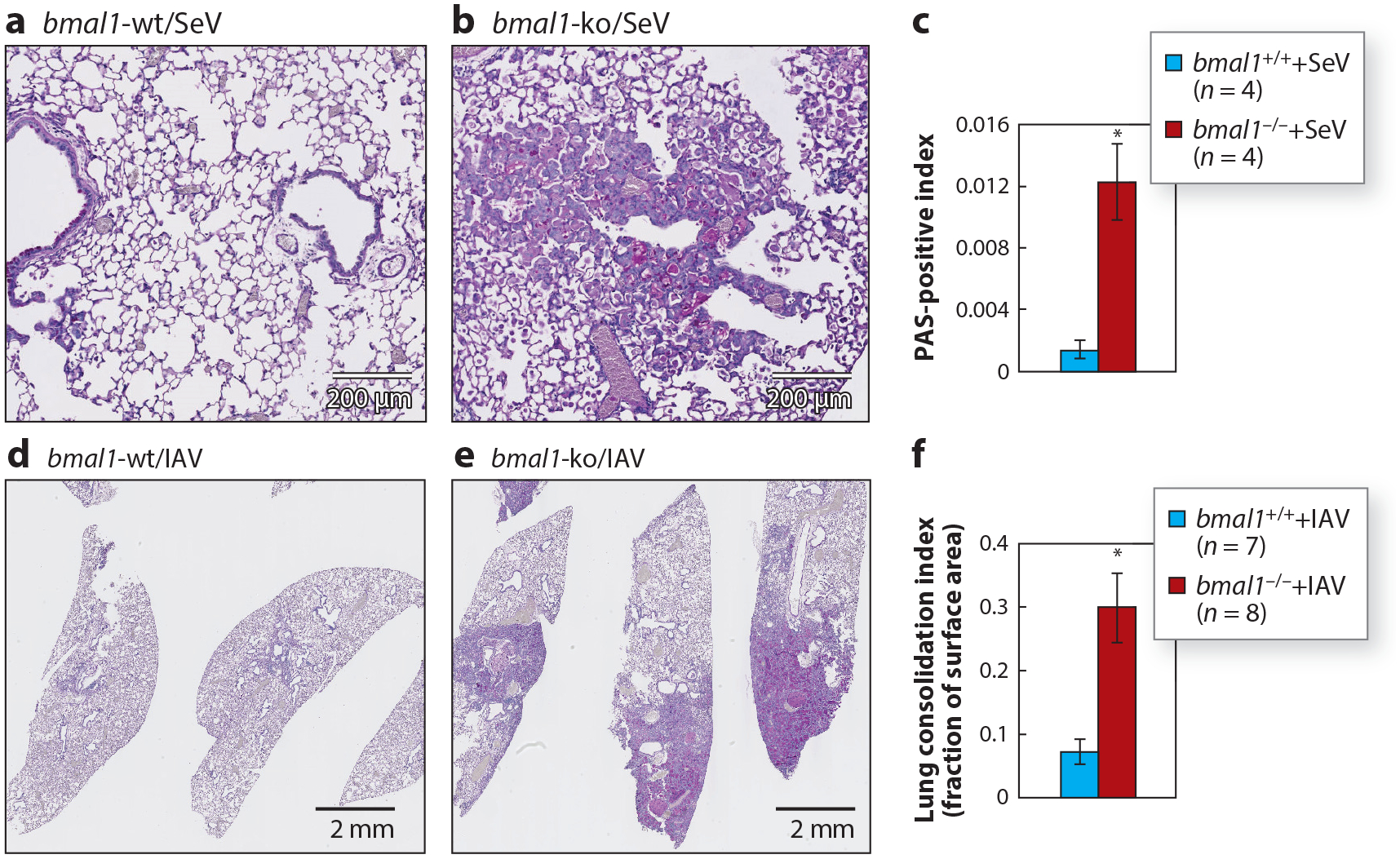
Clock gene bmal1 regulates post-viral chronic lung disease in mice. Mice were intranasally infected with 105 pfu of mouse parainfluenza virus (SeV; a–c) or 5 pfu of influenza virus [influenza A virus (IAV) strain A/WS/33 H1N1; d–f] and PAS-stained histological sections were prepared 49 days or 21 days postinfection, respectively. (a,b) SeV-infected wild-type (wt) and bmal1-knockout (ko) mouse lungs, respectively, as described in our previous report (114). (d,e) IAV-infected mice wt and bmal1-ko mouse lungs, respectively. Morphometric quantification of lung remodeling after SeV or IAV infection based on bmal1 genotype (mean ± SE) is provided in panels c and f, respectively (*p < 0.05, student’s 2-tailed t-test).
Lung Cancer
Environmental disruption of circadian rhythms through chronic shift work is statistically associated with a variety of cancers, and small-molecule REV-ERB agonists were recently shown to have general antitumor activity (135, 136). Focusing on immunologic circadian rhythms within the lung, recent evidence suggests a role for clocks in the control of metastatic tumor burden. Subjecting rats to chronic jet lag increased lung metastatic burden in a model of mammary cell adenocarcinoma, an effect attributed to disruptions in the normal rhythms of natural killer cell cytolytic activity (137). In a melanoma model, lung metastatic tumor burden exhibited a circadian rhythm based on the time that the cancer cells were administered to mice (111). Importantly, these rhythms could be eliminated by deleting bmal1 in neutrophils (111). Taken together, circadian clock function within at least two innate immune effector cells appears to be relevant for the establishment of cancer within the lung, although the specific mechanisms involved appear to be context specific. At present, it is unknown whether circadian clocks intrinsic to adaptive immune cells play a role in lung cancer. However, a recent article implicated bmal1 in the downregulation of programmed death-ligand 1 (PD-L1) on macrophages, an important immune checkpoint molecule (138). Deleting bmal1 in myeloid cells led to increased PD-L1 levels and promoted immune exhaustion in a mouse model of bacterial sepsis (138). Extrapolating these results to cancer, it is tempting to speculate that circadian regulation could be exploited to enhance the efficacy of immunotherapy, especially in lung cancers where PD-L1 expression is known to be variable (139).
FUTURE DIRECTIONS
Over the last five years we have begun to understand how the respiratory system uses temporal information generated by the circadian clock to organize homeostatic immune responses. Immune functions tied to circadian rhythms in the lung to date include the initial detection of pathogens or tumor cells, the initiation of innate immune responses though inflammation, and the orderly control of leukocyte trafficking. Lung circadian rhythms in all their diversity appear to be a tale of many clocks, and specific contributions have now been tied to local clock activity within airway epithelial cells, endothelial cells, and various leukocyte clocks. For all of the progress made on this topic in the last half decade, unknowns still far outweigh the knowns. We know from clinical experience that acquired immune reactions such as allergies have strong daily rhythms in humans, as do vaccine responses (11, 140), but thus far we know little about how circadian clocks or specific clock proteins within the adaptive immune system might contribute to this. The lung contains a great diversity of cell types that are physiologically and clinically important but whose circadian organization remains unexplored, for example, type II pneumocytes, alveolar macrophages, and airway smooth muscle. What we know so far about circadian rhythms in different lung cell types suggests they may be interdependent to a degree. As such, it must be remembered that the usual practice of conditionally knocking out clock genes in a specific cell type has the potential to obscure as well as to clarify. We may need the application of new experimental approaches, such as single-cell expression profiling, to deconvolute the myriad circadian programs at work within the lung. At present, this technology provides a reading depth in the range of thousands of reads per cell, while high-fidelity circadian rhythm detection requires on the order of 20 million reads (141), so additional technical development will be required. Finally, while we now have detailed information on lung circadian gene expression in healthy mice and even humans, we know very little about the temporal programs in chronic lung disease. We also have little information on how chronological age affects circadian gene expression programs. We suspect that the chronically diseased lung may use temporal information generated by circadian clocks very differently and that age is also likely to be an important factor. Understanding the temporal architecture unique to lung disease represents an important path for further study and one that promises to yield valuable insights into respiratory disease and overall human health.
ACKNOWLEDGMENTS
We thank Robyn Haspel and Steven Brody for their editorial input. This work was funded by RO1HL135846 and a Children’s Development Institute grant (PD-II-2016-529).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Karamanou M, Androutsos G. 2011. Aretaeus of Cappadocia and the first clinical description of asthma. Am. J. Respir. Crit. Care Med 184:1420–21 [DOI] [PubMed] [Google Scholar]
- 2.Aretaeus. 1861. The Extant Works of Aretaeus, the Cappadocian, transl. F Adams, London: Sydenham Soc. [Google Scholar]
- 3.Natl. Asthma Educ. Prev. Prog 2007. Guidelines for the diagnosis and management of asthma. Expert Panel Rep. 3, US Dep. Health Hum. Serv., Washington, DC. https://www.nhlbi.nih.gov/files/docs/guidelines/asthsumm.pdf
- 4.Martin RJ. 1992. Circadian rhythms, nocturnal asthma, and management. Ann. Allergy 69:267–72 [PubMed] [Google Scholar]
- 5.Smolensky MH, Barnes PJ, Reinberg A, McGovern JP. 1986. Chronobiology and asthma. I. Day-night differences in bronchial patency and dyspnea and circadian rhythm dependencies. J. Asthma 23:321–43 [DOI] [PubMed] [Google Scholar]
- 6.Refinetti R 2012. Integration of biological clocks and rhythms. Compr. Physiol 2:1213–39 [DOI] [PubMed] [Google Scholar]
- 7.Beaver LM, Gvakharia BO, Vollintine TS, Hege DM, Stanewsky R, Giebultowicz JM. 2002. Loss of circadian clock function decreases reproductive fitness in males of Drosophila melanogaster. PNAS 99:2134–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spoelstra K, Wikelski M, Daan S, Loudon AS, Hau M. 2016. Natural selection against a circadian clock gene mutation in mice. PNAS 113:686–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. 1998. Resonating circadian clocks enhance fitness in cyanobacteria. PNAS 95:8660–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silver AC, Arjona A, Walker WE, Fikrig E. 2012. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity 36:251–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long JE, Drayson MT, Taylor AE, Toellner KM, Lord JM, Phillips AC. 2016. Morning vaccination enhances antibody response over afternoon vaccination: a cluster-randomised trial. Vaccine 34:2679–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortier EE, Rooney J, Dardente H, Hardy MP, Labrecque N, Cermakian N. 2011. Circadian variation of the response of T cells to antigen. J. Immunol 187:6291–300 [DOI] [PubMed] [Google Scholar]
- 13.Bahrami-Nejad Z, Zhao ML, Tholen S, Hunerdosse D, Tkach KE, et al. 2018. A transcriptional circuit filters oscillating circadian hormonal inputs to regulate fat cell differentiation. Cell Metab. 27:854–68.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang G, Chen L, Grant GR, Paschos G, Song WL, et al. 2016. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci. Transl. Med 8:324ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amano H, Fukuda Y, Yokoo T, Yamaoka K. 2018. Interleukin-6 level among shift and night workers in Japan: cross-sectional analysis of the J-HOPE study. J. Atheroscler. Thromb 25:1206–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puttonen S, Viitasalo K, Härmä M. 2011. Effect of shiftwork on systemic markers of inflammation. Chronobiol. Int 28:528–35 [DOI] [PubMed] [Google Scholar]
- 17.Kecklund G, Axelsson J. 2016. Health consequences of shift work and insufficient sleep. BMJ 355:i5210. [DOI] [PubMed] [Google Scholar]
- 18.Papagiannakopoulos T, Bauer MR, Davidson SM, Heimann M, Subbaraj L, et al. 2016. Circadian rhythm disruption promotes lung tumorigenesis. Cell Metab. 24:324–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rensing L, Meyer-Grahle U, Ruoff P. 2001. Biological timing and the clock metaphor: oscillatory and hourglass mechanisms. Chronobiol. Int 18:329–69 [DOI] [PubMed] [Google Scholar]
- 20.Reinberg A, Smolensky MH. 1983. Biological Rhythms and Medicine: Cellular, Metabolic, Physiopathologic, and Pharmacologic Aspects. New York: Springer [Google Scholar]
- 21.Koukkari W, Sothern R. 2006. Introducing Biological Rhythms: A Primer on the Temporal Organization of Life, with Implications for Health, Society, Reproduction, and the Natural Environment. New York: Springer Sci. Bus. [Google Scholar]
- 22.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, et al. 2014. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159:514–29 [DOI] [PubMed] [Google Scholar]
- 23.Adamovich Y, Ladeuix B, Sobel J, Manella G, Neufeld-Cohen A, et al. 2019. Oxygen and carbon dioxide rhythms are circadian clock controlled and differentially directed by behavioral signals. Cell Metab. 29:1092–1103.e3 [DOI] [PubMed] [Google Scholar]
- 24.Spengler CM, Shea SA. 2000. Endogenous circadian rhythm of pulmonary function in healthy humans. Am. J. Respir. Crit. Care Med 162:1038–46 [DOI] [PubMed] [Google Scholar]
- 25.Clark TJ. 1987. Diurnal rhythm of asthma. Chest 91:137S–41S [DOI] [PubMed] [Google Scholar]
- 26.Ballard RD, Saathoff MC, Patel DK, Kelly PL, Martin RJ. 1989. Effect of sleep on nocturnal bronchoconstriction and ventilatory patterns in asthmatics. J. Appl. Physiol 67:243–49 [DOI] [PubMed] [Google Scholar]
- 27.Catterall JR, Rhind GB, Stewart IC, Whyte KF, Shapiro CM, Douglas NJ. 1986. Effect of sleep deprivation on overnight bronchoconstriction in nocturnal asthma. Thorax 41:676–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casale R, Pasqualetti P. 1997. Cosinor analysis of circadian peak expiratory flow variability in normal subjects, passive smokers, heavy smokers, patients with chronic obstructive pulmonary disease and patients with interstitial lung disease. Respiration 64:251–56 [DOI] [PubMed] [Google Scholar]
- 29.Casale R, Natali G, Colantonio D, Pasqualetti P. 1992. Circadian rhythm of peak expiratory flow in children passively exposed and not exposed to cigarette smoke. Thorax 47:801–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spengler CM, Czeisler CA, Shea SA. 2000. An endogenous circadian rhythm of respiratory control in humans. J. Physiol 526(Part 3):683–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler MP, Smales C, Wu H, Hussain MV, Mohamed YA, et al. 2015. The circadian system contributes to apnea lengthening across the night in obstructive sleep apnea. Sleep 38:1793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kondo S 1993. Circadian variation of bronchial caliber and antigen-induced late asthmatic response. Chest 104:801–5 [DOI] [PubMed] [Google Scholar]
- 33.Mohiuddin AA, Martin RJ. 1990. Circadian basis of the late asthmatic response. Am. Rev. Respir. Dis 142:1153–57 [DOI] [PubMed] [Google Scholar]
- 34.Ferraz E, Borges MC, Terra-Filho J, Martinez JA, Vianna EO. 2006. Comparison of 4 am and 4 pm bronchial responsiveness to hypertonic saline in asthma. Lung 184:341–46 [DOI] [PubMed] [Google Scholar]
- 35.Oosterhoff Y, Koeter GH, De Monchy JG, Postma DS. 1993. Circadian variation in airway responsiveness to methacholine, propranolol, and AMP in atopic asthmatic subjects. Am. Rev. Respir. Dis 147:512–17 [DOI] [PubMed] [Google Scholar]
- 36.Davies RJ, Green M, Schofield NM. 1976. Recurrent nocturnal asthma after exposure to grain dust. Am. Rev. Respir. Dis 114:1011–19 [DOI] [PubMed] [Google Scholar]
- 37.Kelly EA, Houtman JJ, Jarjour NN. 2004. Inflammatory changes associated with circadian variation in pulmonary function in subjects with mild asthma. Clin. Exp. Allergy 34:227–33 [DOI] [PubMed] [Google Scholar]
- 38.Huang RC. 2018. The discoveries of molecular mechanisms for the circadian rhythm: the 2017 Nobel Prize in Physiology or Medicine. Biomed. J 41:5–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mackey SR. 2007. Biological Rhythms Workshop IA: molecular basis of rhythms generation. Cold Spring Harb. Symp. Quant. Biol 72:7–19 [DOI] [PubMed] [Google Scholar]
- 40.Trott AJ, Menet JS. 2018. Regulation of circadian clock transcriptional output by CLOCK:BMAL1. PLOS Genet. 14:e1007156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Yang G. 2014. PPARs integrate the mammalian clock and energy metabolism. PPAR Res. 2014:653017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. 2006. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 4:25–36 [DOI] [PubMed] [Google Scholar]
- 43.Storch KF, Paz C, Signorovitch J, Raviola E, Pawlyk B, et al. 2007. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell 130:730–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patke A, Murphy PJ, Onat OE, Krieger AC, Ozcelik T, et al. 2017. Mutation of the human circadian clock gene CRY1 in familial delayed sleep phase disorder. Cell 169:203–15.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, et al. 2001. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291:1040–43 [DOI] [PubMed] [Google Scholar]
- 46.Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, et al. 2005. Functional consequences of a CKIδ mutation causing familial advanced sleep phase syndrome. Nature 434:640–44 [DOI] [PubMed] [Google Scholar]
- 47.Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. 2008. Redundant function of REV-ERBα and β and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLOS Genet. 4:e1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, et al. 2005. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308:414–15 [DOI] [PubMed] [Google Scholar]
- 49.O’Neill JS, Reddy AB. 2011. Circadian clocks in human red blood cells. Nature 469:498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scrima R, Cela O, Merla G, Augello B, Rubino R, et al. 2016. Clock-genes and mitochondrial respiratory activity: evidence of a reciprocal interplay. Biochim. Biophys. Acta 1857:1344–51 [DOI] [PubMed] [Google Scholar]
- 51.Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, et al. 2009. Harmonics of circadian gene transcription in mammals. PLOS Genet. 5:e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perrin L, Loizides-Mangold U, Chanon S, Gobet C, Hulo N, et al. 2018. Transcriptomic analyses reveal rhythmic and CLOCK-driven pathways in human skeletal muscle. eLife 7:e34114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, et al. 2012. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338:349–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Song L, Liu M, Ge R, Zhou Q, et al. 2018. A proteomics landscape of circadian clock in mouse liver. Nat. Commun 9:1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kriebs A, Jordan SD, Soto E, Henriksson E, Sandate CR, et al. 2017. Circadian repressors CRY1 and CRY2 broadly interact with nuclear receptors and modulate transcriptional activity. PNAS 114:8776–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang X, Lamia KA, Evans RM. 2007. Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb. Symp. Quant. Biol 72:387–94 [DOI] [PubMed] [Google Scholar]
- 57.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, et al. 2007. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat. Struct. Mol. Biol 14:1207–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burris TP, Busby SA, Griffin PR. 2012. Targeting orphan nuclear receptors for treatment of metabolic diseases and autoimmunity. Chem. Biol 19:51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, et al. 2015. The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell 161:1138–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu R, Dang F, Li P, Wang P, Xu Q, et al. 2019. The circadian protein Period2 suppresses mTORC1 activity via recruiting Tsc1 to mTORC1 complex. Cell Metab. 29:653–67.e6 [DOI] [PubMed] [Google Scholar]
- 61.Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, et al. 2010. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat. Med 16:1152–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, et al. 2015. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348:1488–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okabe T, Chavan R, Fonseca Costa SS, Brenna A, Ripperger JA, Albrecht U. 2016. REV-ERBα influences the stability and nuclear localization of the glucocorticoid receptor. J. Cell Sci 129:4143–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee Y, Kim K. 2012. Posttranslational and epigenetic regulation of the CLOCK/BMAL1 complex in the mammalian. Anim. Cells Syst 16:1–10 [Google Scholar]
- 65.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, et al. 2004. PERIOD2::LUCIFERASE realtime reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. PNAS 101:5339–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. 2004. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119:693–705 [DOI] [PubMed] [Google Scholar]
- 67.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. 2004. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol 14:2289–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schibler U, Gotic I, Saini C, Gos P, Curie T, et al. 2015. Clock-talk: interactions between central and peripheral circadian oscillators in mammals. Cold Spring Harb. Symp. Quant. Biol 80:223–32 [DOI] [PubMed] [Google Scholar]
- 69.Lehman MN, Silver R, Gladstone WR, Kahn RM, Gibson M, Bittman EL. 1987. Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J. Neurosci 7:1626–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hughes ME, Hong HK, Chong JL, Indacochea AA, Lee SS, et al. 2012. Brain-specific rescue of Clock reveals system-driven transcriptional rhythms in peripheral tissue. PLOS Genet. 8:e1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. 2007. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLOS Biol. 5:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Z, Hunter L, Wu G, Maidstone R, Mizoro Y, et al. 2019. Genome-wide effect of pulmonary airway epithelial cell-specific Bmal1 deletion. FASEB J. 33:6226–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruben MD, Wu G, Smith DF, Schmidt RE, Francey LJ, et al. 2018. A database of tissue-specific rhythmically expressed human genes has potential applications in circadian medicine. Sci. Transl. Med 10:eaat8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anafi RC, Francey LJ, Hogenesch JB, Kim J. 2017. CYCLOPS reveals human transcriptional rhythms in health and disease. PNAS 114:5312–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. 2014. A circadian gene expression atlas in mammals: implications for biology and medicine. PNAS 111:16219–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, et al. 2018. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 359:eaao0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mavroudis PD, DuBois DC, Almon RR, Jusko WJ. 2018. Daily variation of gene expression in diverse rat tissues. PLOS ONE 13:e0197258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beytebiere JR, Trott AJ, Greenwell BJ, Osborne CA, Vitet H, et al. 2019. Tissue-specific BMAL1 cistromes reveal that rhythmic transcription is associated with rhythmic enhancer-enhancer interactions. Genes Dev. 33:294–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caratti G, Iqbal M, Hunter L, Kim D, Wang P, et al. 2018. REVERBα couples the circadian clock to hepatic glucocorticoid action. J. Clin. Investig 128:4454–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haspel JA, Chettimada S, Shaik RS, Chu JH, Raby BA, et al. 2014. Circadian rhythm reprogramming during lung inflammation. Nat. Commun 5:4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramasamy A, Kuokkanen M, Vedantam S, Gajdos ZK, Couto Alves A, et al. 2012. Genome-wide association studies of asthma in population-based cohorts confirm known and suggested loci and identify an additional association near HLA. PLOS ONE 7:e44008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, et al. 2010. A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med 363:1211–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Acevedo N, Saaf A, Soderhall C, Melen E, Mandelin J, et al. 2013. Interaction between retinoid acid receptor-related orphan receptor alpha (RORA) and neuropeptide S receptor 1 (NPSR1) in asthma. PLOS ONE 8:e60111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Franks TJ, Colby TV, Travis WD, Tuder RM, Reynolds HY, et al. 2008. Resident cellular components of the human lung: current knowledge and goals for research on cell phenotyping and function. Proc. Am. Thorac. Soc 5:763–66 [DOI] [PubMed] [Google Scholar]
- 85.Ermert L, Duncker HR, Rosseau S, Schutte H, Seeger W. 1994. Morphometric analysis of pulmonary intracapillary leukocyte pools in ex vivo-perfused rabbit lungs. Am. J. Physiol 267:L64–70 [DOI] [PubMed] [Google Scholar]
- 86.Granton E, Kim JH, Podstawka J, Yipp BG. 2018. The lung microvasculature is a functional immune niche. Trends Immunol. 39:890–99 [DOI] [PubMed] [Google Scholar]
- 87.Pezuk P, Mohawk JA, Wang LA, Menaker M. 2012. Glucocorticoids as entraining signals for peripheral circadian oscillators. Endocrinology 153:4775–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Buhr ED, Yoo SH, Takahashi JS. 2010. Temperature as a universal resetting cue for mammalian circadian oscillators. Science 330:379–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bando H, Nishio T, van der Horst GT, Masubuchi S, Hisa Y, Okamura H. 2007. Vagal regulation of respiratory clocks in mice. J. Neurosci 27:4359–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gibbs J, Ince L, Matthews L, Mei J, Bell T, et al. 2014. An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat. Med 20:919–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ince LM, Zhang Z, Beesley S, Vonslow RM, Saer BR, et al. 2019. Circadian variation in pulmonary inflammatory responses is independent of rhythmic glucocorticoid signaling in airway epithelial cells. FASEB J. 33:126–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Abraham U, Granada AE, Westermark PO, Heine M, Kramer A, Herzel H. 2010. Coupling governs entrainment range of circadian clocks. Mol. Syst. Biol 6:438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. 2002. Effects of aging on central and peripheral mammalian clocks. PNAS 99:10801–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, et al. 2019. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med 199:1517–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, et al. 2018. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560:319–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Williams J, Yang N, Wood A, Zindy E, Meng QJ, Streuli CH. 2018. Epithelial and stromal circadian clocks are inversely regulated by their mechano-matrix environment. J. Cell Sci 131:jcs208223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gibbs JE, Beesley S, Plumb J, Singh D, Farrow S, et al. 2009. Circadian timing in the lung; a specific role for bronchiolar epithelial cells. Endocrinology 150:268–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Du Y, Kitzmiller JA, Sridharan A, Perl AK, Bridges JP, et al. 2017. Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development. Thorax 72:481–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pariollaud M, Gibbs JE, Hopwood TW, Brown S, Begley N, et al. 2018. Circadian clock component REV-ERBα controls homeostatic regulation of pulmonary inflammation. J. Clin. Investig 128:2281–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Halberg F, Johnson EA, Brown BW, Bittner JJ. 1960. Susceptibility rhythm to E. coli endotoxin and bioassay. Proc. Soc. Exp. Biol. Med 103:142–44 [DOI] [PubMed] [Google Scholar]
- 101.Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, et al. 2012. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. PNAS 109:582–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ryzhikov M, Ehlers A, Steinberg D, Xie W, Oberlander E, et al. 2019. Diurnal rhythms spatially and temporally organize autophagy. Cell Rep. 26:1880–92.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oishi K, Ohkura N, Kadota K, Kasamatsu M, Shibusawa K, et al. 2006. Clock mutation affects circadian regulation of circulating blood cells. J. Circadian Rhythms 4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Scheiermann C, Kunisaki Y, Lucas D, Chow A, Jang JE, et al. 2012. Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity 37:290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao Y, Liu M, Chan XY, Tan SY, Subramaniam S, et al. 2017. Uncovering the mystery of opposite circadian rhythms between mouse and human leukocytes in humanized mice. Blood 130:1995–2005 [DOI] [PubMed] [Google Scholar]
- 106.Casanova-Acebes M, Pitaval C, Weiss LA, Nombela-Arrieta C, Chevre R, et al. 2013. Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell 153:1025–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.He W, Holtkamp S, Hergenhan SM, Kraus K, de Juan A, et al. 2018. Circadian expression of migratory factors establishes lineage-specific signatures that guide the homing of leukocyte subsets to tissues. Immunity 49:1175–90.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.A-Gonzalez N, Quintana JA, García-Silva S, Mazariegos M, González de la Aleja A, et al. 2017. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J. Exp. Med 214:1281–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Druzd D, Matveeva O, Ince L, Harrison U, He W, et al. 2017. Lymphocyte circadian clocks control lymph node trafficking and adaptive immune responses. Immunity 46:120–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Adrover JM, Del Fresno C, Crainiciuc G, Cuartero MI, Casanova-Acebes M, et al. 2019. A neutrophil timer coordinates immune defense and vascular protection. Immunity 50:390–402.e10 [DOI] [PubMed] [Google Scholar]
- 111.Casanova-Acebes M, Nicolas-Avila JA, Li JL, Garcia-Silva S, Balachander A, et al. 2018. Neutrophils instruct homeostatic and pathological states in naive tissues. J. Exp. Med 215:2778–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hriscu ML. 2005. Modulatory factors of circadian phagocytic activity. Ann. N. Y. Acad. Sci 1057:403–30 [DOI] [PubMed] [Google Scholar]
- 113.Pavord ID, Beasley R, Agusti A, Anderson GP, Bel E, et al. 2018. After asthma: redefining airways diseases. Lancet 391:350–400 [DOI] [PubMed] [Google Scholar]
- 114.Ehlers A, Xie W, Agapov E, Brown S, Steinberg D, et al. 2018. BMAL1 links the circadian clock to viral airway pathology and asthma phenotypes. Mucosal Immunol. 11:97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Majumdar T, Dhar J, Patel S, Kondratov R, Barik S. 2017. Circadian transcription factor BMAL1 regulates innate immunity against select RNA viruses. Innate Immunity 23:147–54 [DOI] [PubMed] [Google Scholar]
- 116.Zaslona Z, Case S, Early JO, Lalor SJ, McLoughlin RM, et al. 2017. The circadian protein BMAL1 in myeloid cells is a negative regulator of allergic asthma. Am. J. Physiol. Lung Cell. Mol. Physiol 312:L855–60 [DOI] [PubMed] [Google Scholar]
- 117.Falliers CJ. 1981. Time-dependent processes in allergy and asthma. Ann. Allergy 47:253–59 [PubMed] [Google Scholar]
- 118.Gervais P, Reinberg A, Gervais C, Smolensky M, DeFrance O. 1977. Twenty-four-hour rhythm in the bronchial hyperreactivity to house dust in asthmatics. J. Allergy Clin. Immunol 59:207–13 [DOI] [PubMed] [Google Scholar]
- 119.Wenzel S, Holgate ST. 2006. The mouse trap: it still yields few answers in asthma. Am. J. Respir. Crit. Care Med 174:1173–76 [DOI] [PubMed] [Google Scholar]
- 120.Meltzer LJ, Faino A, Szefler SJ, Strand M, Gelfand EW, Beebe DW. 2015. Experimentally manipulated sleep duration in adolescents with asthma: feasibility and preliminary findings. Pediatr. Pulmonol 50:1360–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Noonan M, Karpel JP, Bensch GW, Ramsdell JW, Webb DR, et al. 2001. Comparison of once-daily to twice-daily treatment with mometasone furoate dry powder inhaler. Ann. Allergy Asthma Immunol 86:36–43 [DOI] [PubMed] [Google Scholar]
- 122.Tsai CL, Brenner BE, Camargo CA Jr. 2007. Circadian-rhythm differences among emergency department patients with chronic obstructive pulmonary disease exacerbation. Chronobiol. Int 24:699–713 [DOI] [PubMed] [Google Scholar]
- 123.Hwang JW, Sundar IK, Yao H, Sellix MT, Rahman I. 2014. Circadian clock function is disrupted by environmental tobacco/cigarette smoke, leading to lung inflammation and injury via a SIRT1-BMAL1 pathway. FASEB J. 28:176–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sundar IK, Ahmad T, Yao H, Hwang JW, Gerloff J, et al. 2015. Influenza A virus-dependent remodeling of pulmonary clock function in a mouse model of COPD. Sci. Rep 4:9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lederer DJ, Martinez FJ. 2018. Idiopathic pulmonary fibrosis. N. Engl. J. Med 378:1811–23 [DOI] [PubMed] [Google Scholar]
- 126.Nureki SI, Tomer Y, Venosa A, Katzen J, Russo SJ, et al. 2018. Expression of mutant Sftpc in murine alveolar epithelia drives spontaneous lung fibrosis. J. Clin. Investig 128:4008–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mora AL, Bueno M, Rojas M. 2017. Mitochondria in the spotlight of aging and idiopathic pulmonary fibrosis. J. Clin. Investig 127:405–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dik WA, McAnulty RJ, Versnel MA, Naber BA, Zimmermann LJ, et al. 2003. Short course dexamethasone treatment following injury inhibits bleomycin induced fibrosis in rats. Thorax 58:765–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pekovic-Vaughan V, Gibbs J, Yoshitane H, Yang N, Pathiranage D, et al. 2014. The circadian clock regulates rhythmic activation of the NRF2/glutathione-mediated antioxidant defense pathway to modulate pulmonary fibrosis. Genes Dev. 28:548–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dong C, Gongora R, Sosulski ML, Luo F, Sanchez CG. 2016. Regulation of transforming growth factorbeta1 (TGF-β1)-induced pro-fibrotic activities by circadian clock gene BMAL1. Respirat. Res 17:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sengupta S, Tang SY, Devine J, Nayak S, Zhang S, et al. 2019. Circadian control of lung inflammation in influenza infection. Nat. Commun 10:4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Edgar RS, Stangherlin A, Nagy AD, Nicoll MP, Efstathiou S, et al. 2016. Cell autonomous regulation of herpes and influenza virus infection by the circadian clock. PNAS 113:10085–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Keeler SP, Agapov EV, Hinojosa ME, Letvin AN, Wu K, Holtzman MJ. 2018. Influenza A virus infection causes chronic lung disease linked to sites of active viral RNA remnants. J. Immunol 201:2354–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Walter MJ, Morton JD, Kajiwara N, Agapov E, Holtzman MJ. 2002. Viral induction of a chronic asthma phenotype and genetic segregation from the acute response. J. Clin. Investig 110:165–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Masri S, Sassone-Corsi P. 2018. The emerging link between cancer, metabolism, and circadian rhythms. Nat. Med 24:1795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sulli G, Rommel A, Wang X, Kolar MJ, Puca F, et al. 2018. Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature 553:351–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Logan RW, Zhang C, Murugan S, O’Connell S, Levitt D, et al. 2012. Chronic shift-lag alters the circadian clock of NK cells and promotes lung cancer growth in rats. J. Immunol 188:2583–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Deng W, Zhu S, Zeng L, Liu J, Kang R, et al. 2018. The circadian clock controls immune checkpoint pathway in sepsis. Cell Rep. 24:366–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yu H, Boyle TA, Zhou C, Rimm DL, Hirsch FR. 2016. PD-L1 expression in lung cancer. J. Thorac. Oncol 11:964–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Smolensky MH, Lemmer B, Reinberg AE. 2007. Chronobiology and chronotherapy of allergic rhinitis and bronchial asthma. Adv. Drug Deliv. Rev 59:852–82 [DOI] [PubMed] [Google Scholar]
- 141.Hughes ME, Abruzzi KC, Allada R, Anafi R, Arpat AB, et al. 2017. Guidelines for genome-scale analysis of biological rhythms. J. Biol. Rhythms 32:380–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, et al. 2002. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 419:841–44 [DOI] [PubMed] [Google Scholar]


