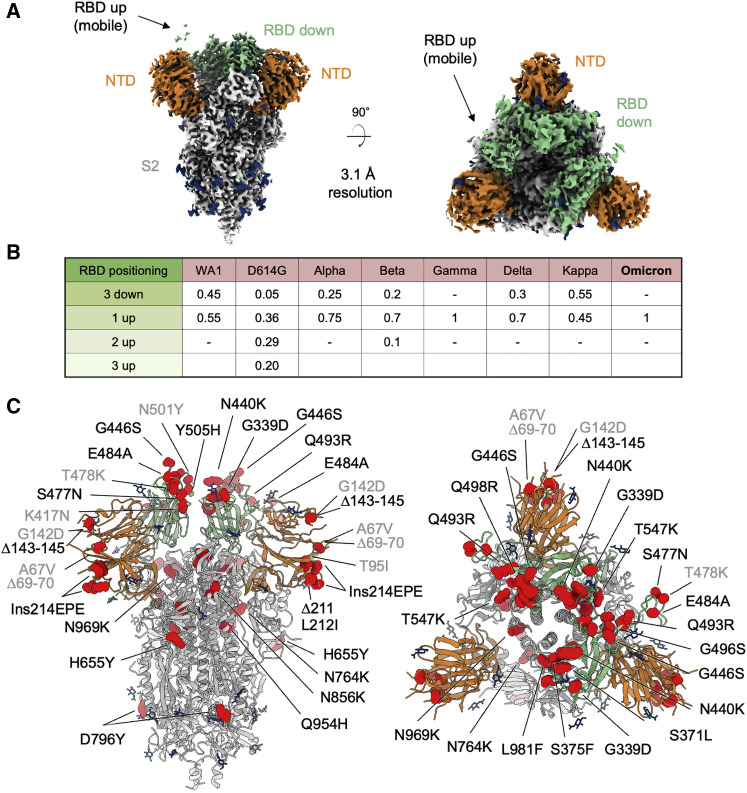
Figure 1.
Cryo-EM structure of prefusion SARS-CoV-2 Omicron (B.1.1.529) spike
(A) Cryo-EM map of SARS-CoV-2 Omicron S2P spike in the prefusion state shown in two orthogonal views. The density for the single RBD up is barely visible at the optimal contour level due to high mobility of the domain. NTD is colored in orange, RBD in green, glycans in blue, the rest of the trimer in gray.
(B) Relative population of RBD states observed in cryo-EM structures of SARS-CoV-2 spike for different variants.
(C) Structure of SARS-CoV-2 Omicron spike in the 1 RBD-up state with mutations highlighted in red. Mutations observed in previous variants are labeled in gray, new Omicron mutations are labeled in black. See also Figures S1, S2, and S3; Tables S1 and S2.