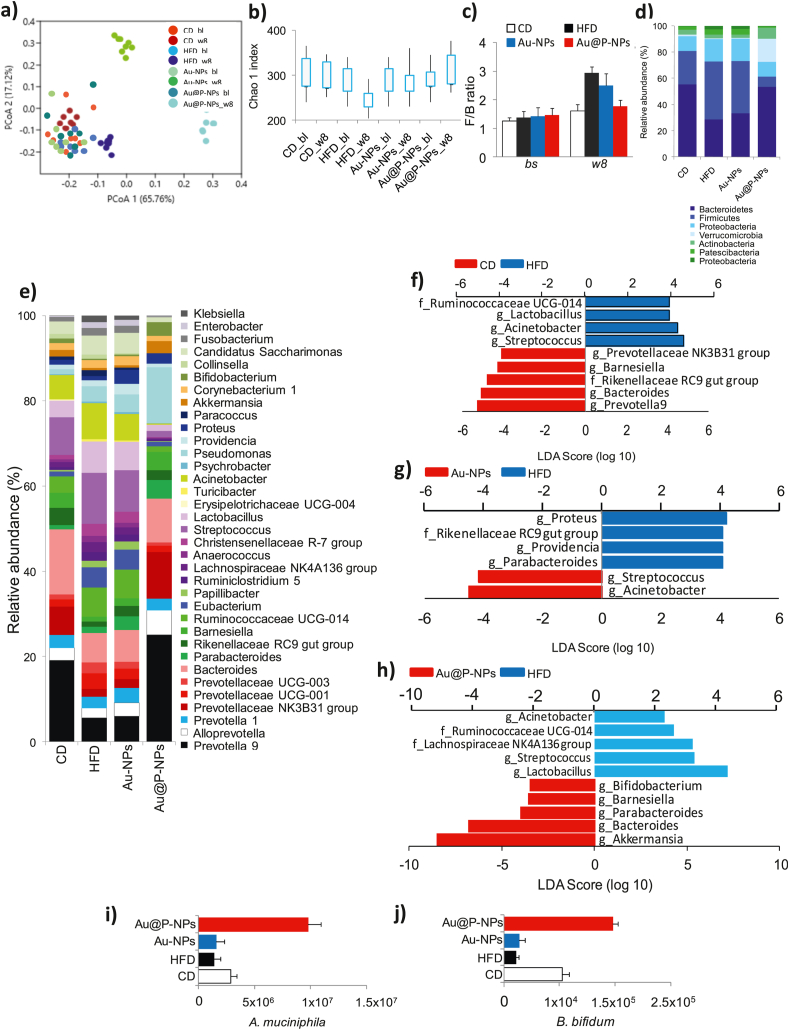
Fig. 5.
Effect of Au@P-NPs on obesity-associated gut microbiota imbalance in HFD-fed mice. At week 8, faecal pellets of indicated treatments were collected. Faecal samples were also harvested during the last week of adaptation on a CD (indicated to bl), before the administration of the HFD or Au@P-NPs. After getting good quality of genomic DNA, 16 S rRNA gene-based gut microbiota profiling was carried out. (a) PCoA. (b) Chao1 index at OTU level. (c) F/B ratio. The relative richness of phylotypes at (d) phylum and (e) lowest taxonomic level (LTL) attained. LEfSe was figured out to investigate the taxa within the LTL likely that more sturdily distinguish between the gut microbiota of (f) CD vs HFD, (g) Au-NPs vs HFD, and (h) Au@P-NPs vs HFD. (i) qPCR amplification of A. muciniphila normalized by 3 × 109 copies of 16 S (total bacteria) in faecal sample. (b–e) KWT with DMCT. (i) One-way frequent measurements ANOVA with a BPHT. ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗P < 0.001 for CD vs HFD; $P < 0.05, $$P < 0.01 and $$$P < 0.001 for Au@P-NPs vs HFD.