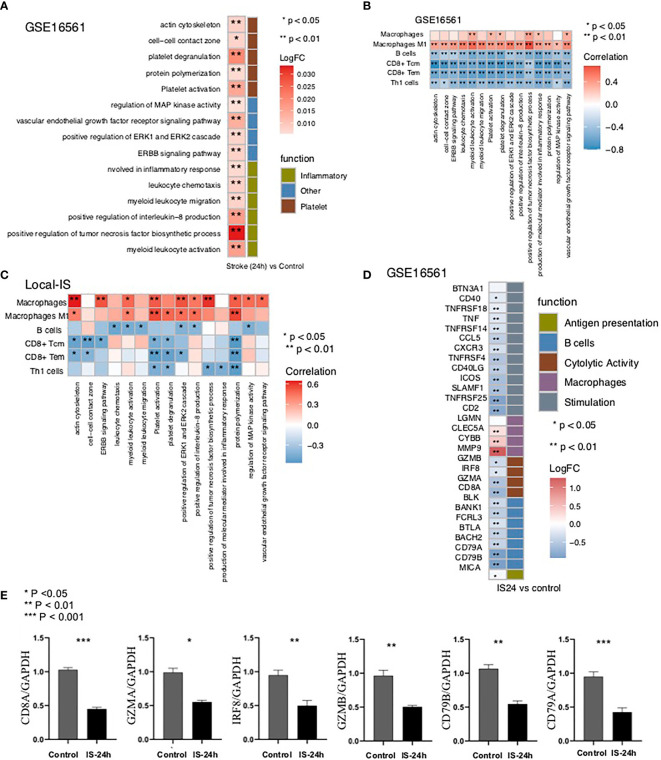
Figure 5.
The result of ssGSEA. (A) Heatmap depicting the mean differences in ssGSEA scores for several signaling pathways between the IS-24h and control groups in GSE16561. The y axis indicates the inflammation-related, platelet-related, and pathological pathways. Each square represents the fold change or difference in the ssGSEA score for an inflammation-related, platelet-related, or pathological pathway between the IS-24h and control groups. Red indicates upregulation, while blue indicates downregulation. (B) Correlation analysis of the scores of different immune cells estimated by the xCell algorithm and the ssGSEA scores for signaling pathways in GSE16561. (C) Correlation analysis of the scores for different immune cells estimated by the xCell algorithm and the ssGSEA scores for signaling pathways in the Local-IS cohort. (D) Heatmap depicting the mean differences in the mRNA expression of immune-related genes between the IS-24h and control groups in GSE16561. The y axis indicates the immune signature or gene compared between the IS-24h and control groups in GSE16561. Each square represents the fold change or difference in each indicated immune signature or gene between the IS-24h and control groups. Red indicates upregulation, while blue indicates downregulation. (E) Expression profile of CD8A, GZMA, IRF8, GZMB, CD79B and CD79A between IS-24h and control groups in Local-IS (qPCR). *P < 0.05; **P < 0.01; ***P < 0.001.