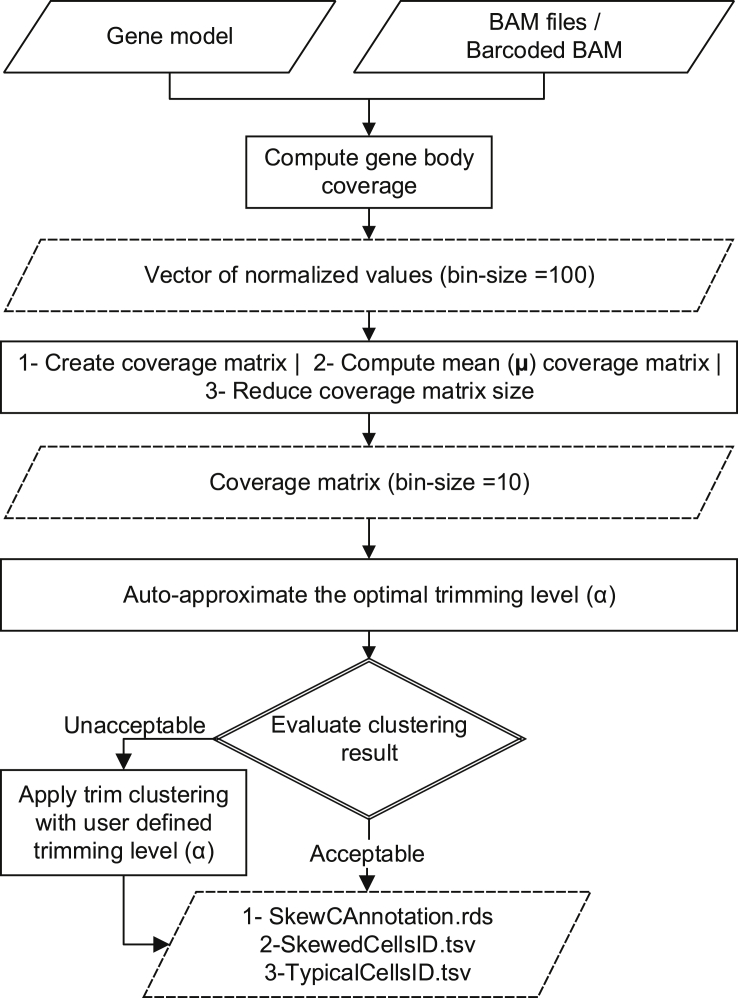
Figure 2.
Overview of SkewC workflow
The figure illustrates the SkewC workflow and implementation to discriminate skewed cells with skewed gene coverage distribution. The circle numbers callout points to the main inputs, processing, and outputs of SkewC. SkewC inputs are the gene model in. bed format and the aligned reads in BAM format per each cell. For scRNA-seq dataset generated by 10x Genomics libraries, the input to SkewC is the postsorted BAM file together with the cell barcode text file. SkewC bash command to supply the inputs (0_split10XbyBarcode.sh) the batch split the postsorted BAM file into individual BAM files. Compute gene body coverage for each cell. SkewC batch script 1_geneBodyCoverage.sh used to compute gene body coverage and produce a text file.r which contains vector of normalized values. The normalized values are stored as a matrix (coverage matrix with bin size = 100), the coverage matrix should be processed by computing the mean of the coverage matrix and reduce the bin size to be 10. The mean coverage matrix used as in put for the batch script 2_SkewC.sh, which use the trimming clustering function in R (tclust) to cluster the coverage matrix, the script designed to auto approximate the optimal trimming level alpha (α) and select the clustering result with optimal alpha (α). Other option is to apply the trim clustering with user-defined trimming level alpha (α). The output provided in different formats, two text files each with the list of the typical and skewed cells. The other format was R data frame object SkewCAnnotation.rds. The list of annotated single-cells can be added to the R Bioconductor SingleCellExperiment Class or Seurat R package object to be used in QC for filtering skewed cells from analysis.