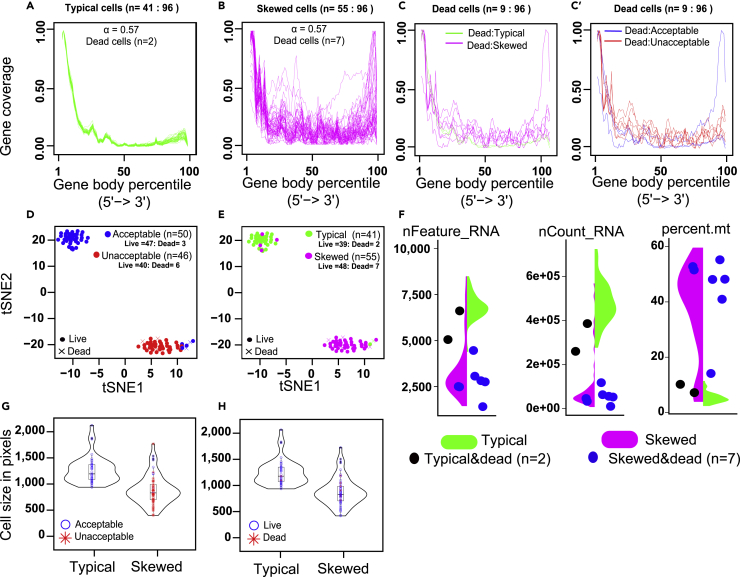
Figure 5.
Validation of SkewC method and biological features of skewed cell using (NCBI GEO: GSE46980)
We used an existing experimentally validated dataset to validate SkewC. The dataset used was (NCBI GEO: GSE46980).
(A–C) the gene body coverage plots for typical, skewed, and all dead cells. The gene body coverage of the dead cells colored based on the SkewC annotation (C) and the (NCBI GEO: GSE46980) authors’ annotation (C′).
(D and E) t-SNE illustrating the clustering of cells as annotated by the (NCBI GEO: GSE46980) dataset authors (Acceptable and Unacceptable) (D) and typical and skewed cells (E). Dead and live cells are highlighted as well in t-SNE.
(F) Violin plots showing Seurat QC metrics. The violin plot split by the SkewC annotation and dead cells are shown as black and blue circles.
(G) The distribution of cell size between typical and skewed cells, acceptable and unacceptable are highlighted.
(H) The distribution of cell size between typical and skewed cells, dead and live is highlighted.