FIGURE 3.
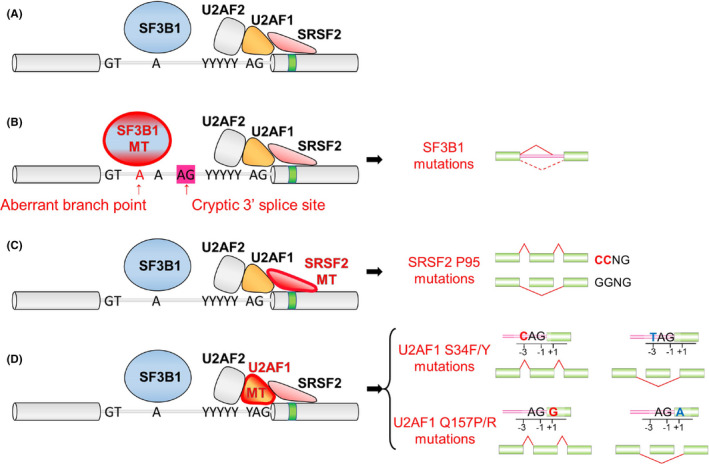
Functional consequences of SF3B1, SRSF2, and U2AF1 mutations in RNA splicing. A, Description of canonical splicing. B, Mutations in SF3B1 (marked in red) result in enhanced usage of an aberrant branchpoint to generate an alternative 3′ss. C, Mutations in SRSF2 are clustered at the proline 95 residue and alter the preference of ESE motif recognition. D, U2AF1 mutations alter the 3′ss consensus sequences. U2AF1 S34F/Y mutations favor inclusion of cassette exons with a C‐nucleotide at the “−3” position, whereas U2AF1 Q157P/R mutations favor inclusion of cassette exons with a G‐nucleotides at the “+1” position
