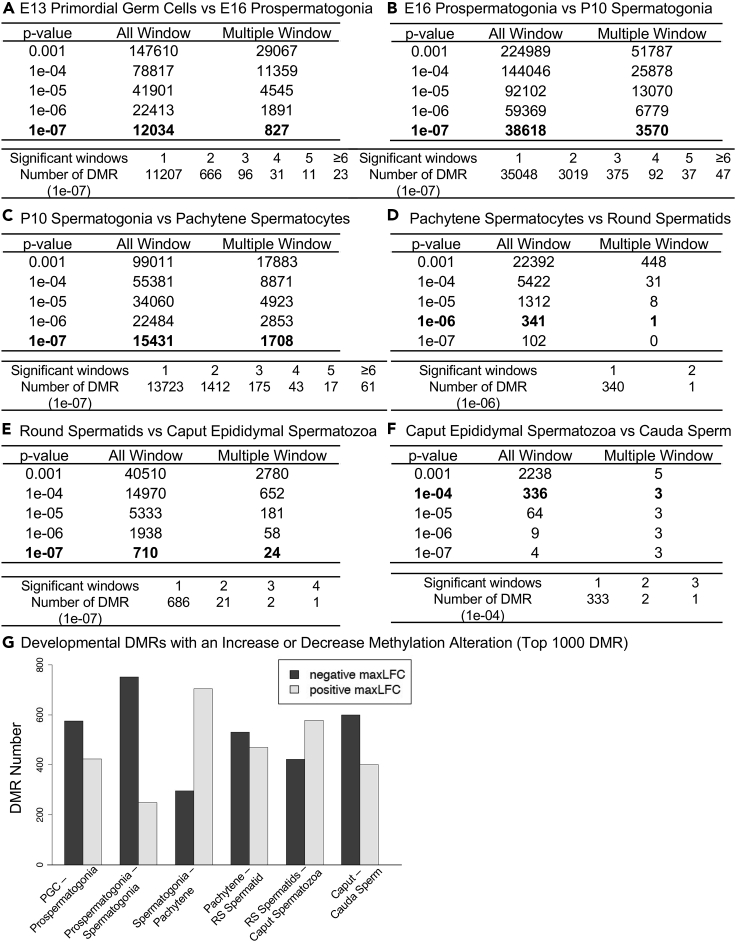
Figure 2.
DMR identification
The number of DMRs found using different p value cutoff thresholds. The All Window column shows all DMR sites. The Multiple Window column shows the number of DMRs with ≥2 significant nearby windows at a p value threshold indicated in the bolded line.
(A) E13 primordial germ cells versus E16 prospermatogonia.
(B) E16 prospermatogonia versus P10 spermatogonia.
(C) P10 spermatogonia versus pachytene spermatocytes.
(D) Pachytene spermatocytes versus round spermatids.
(E) Round spermatids versus caput epididymal spermatozoa.
(F) Caput epididymal spermatozoa versus cauda sperm.
(G) Developmental DMR numbers with an increase or decrease in methylation alteration (top 1000 DMR) from the maximum log-fold-change (maxLFC). Light shading, positive increase in methylation and dark shading, negative decrease in DNA methylation.