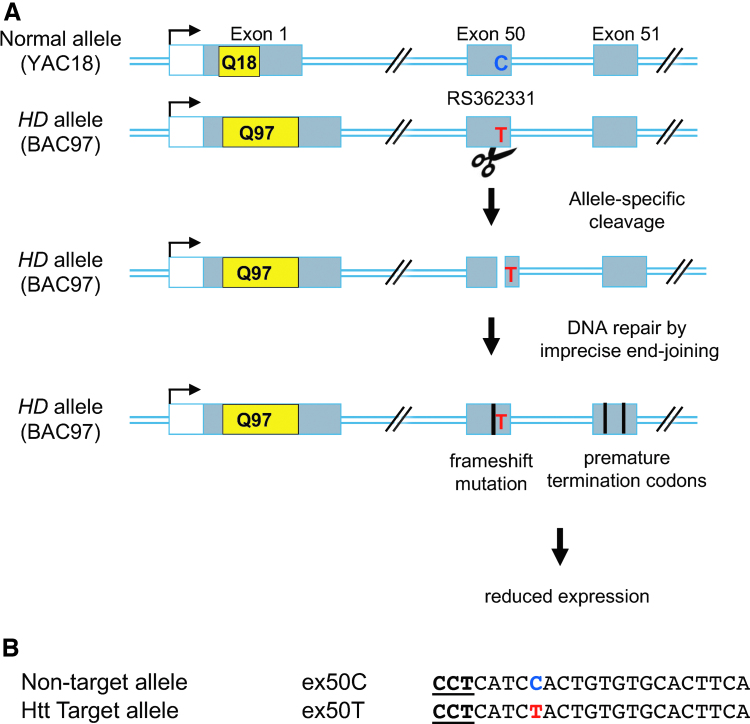
Figure 1.
Strategy for allele-specific targeting of mHTT protein by gene editing. (A) A strategy for allele-specific degradation of mHTT with SNP-specific nucleases. A schematic of the Htt gene depicts 3 of the 67 exons (not drawn to scale). Exon 1 can have a variable number of CAG repeats (indicated in yellow). HD results when a single copy of the gene has a repeat number >36. To specifically alter the disease allele, an allele-specific variable number of trinucleotide (CAG) repeats encoding glutamine (indicated in yellow) nuclease targets an SNP that is heterozygous in an HD individual. In a mouse model of HD, the YAC18 transgene carries a normal version of the human HTT locus with the C allele of SNP RS362331, and the BAC97 transgene carries a mutant human HTT locus and the T allele of SNP RS362331. DNA cleavage and imprecise repair at the mHTT gene result in frameshift mutations that disrupt the coding region of the HD allele. Premature stop codons can reduce expression of the entire HTT protein, possibly through nonsense-mediated decay of the mature mRNA. (B) CRISPR, clustered regularly interspaced short palindromic repeats-Cas9 target sequences specific for two alleles of exon 50 are shown. The heterozygous SNP is shown in blue or red. The PAM sequence is in bold. CAG; CRISPR, clustered regularly interspaced short palindromic repeats; HD, Huntington's disease; HTT, Huntingtin; mHTT, mutant HTT; PAM, protospacer adjacent motif; SNP, single nucleotide polymorphism. Color images are available online.