Fig. 1. Machine learning model based on directed evolution predicts highly active abiotic miniproteins for macromolecule delivery.
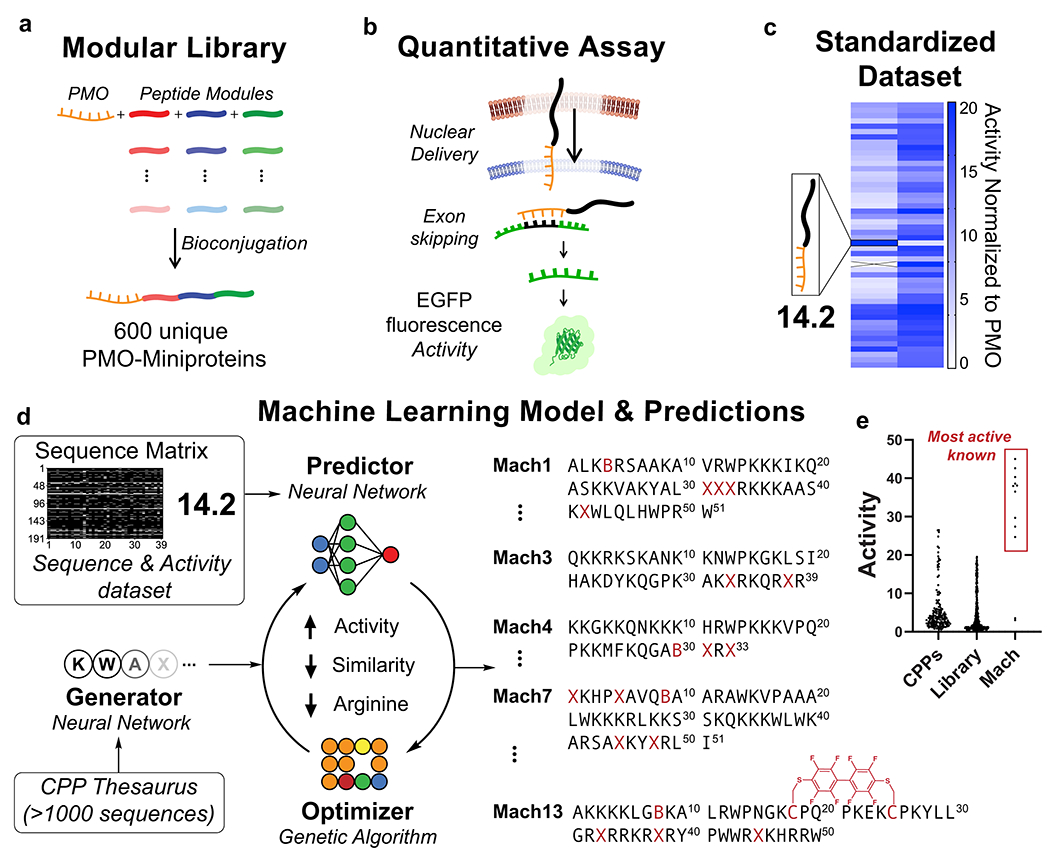
(a) A 600-membered library of PMO-miniprotein conjugates was synthesized using linear combinations of abiotic peptide modules. (b) A standardized in vitro activity assay tests for nuclear delivery using a quantitative fluorescence readout. (c) Members of the modular library exhibit a broad spectrum of activities. Each bit corresponds to a PMO-peptide in the library and its corresponding activity. (d) Sequences are encoded into a fingerprint matrix, labeled with experimental activity, and used to train a machine learning model. The model designs novel sequences in a loop based on directed evolution. X = aminohexanoic acid, B = β-alanine, C = cysteine macrocycles linked through decafluorobiphenyl. (e) Normalized activity from peptides designed in this work (Mach) compared to peptides in the modular library and known CPPs tested using the same assay.
