Extended Data Figure 2. Mutagenesis mediated by type III-A CRISPR-Cas immunity against phage infection.
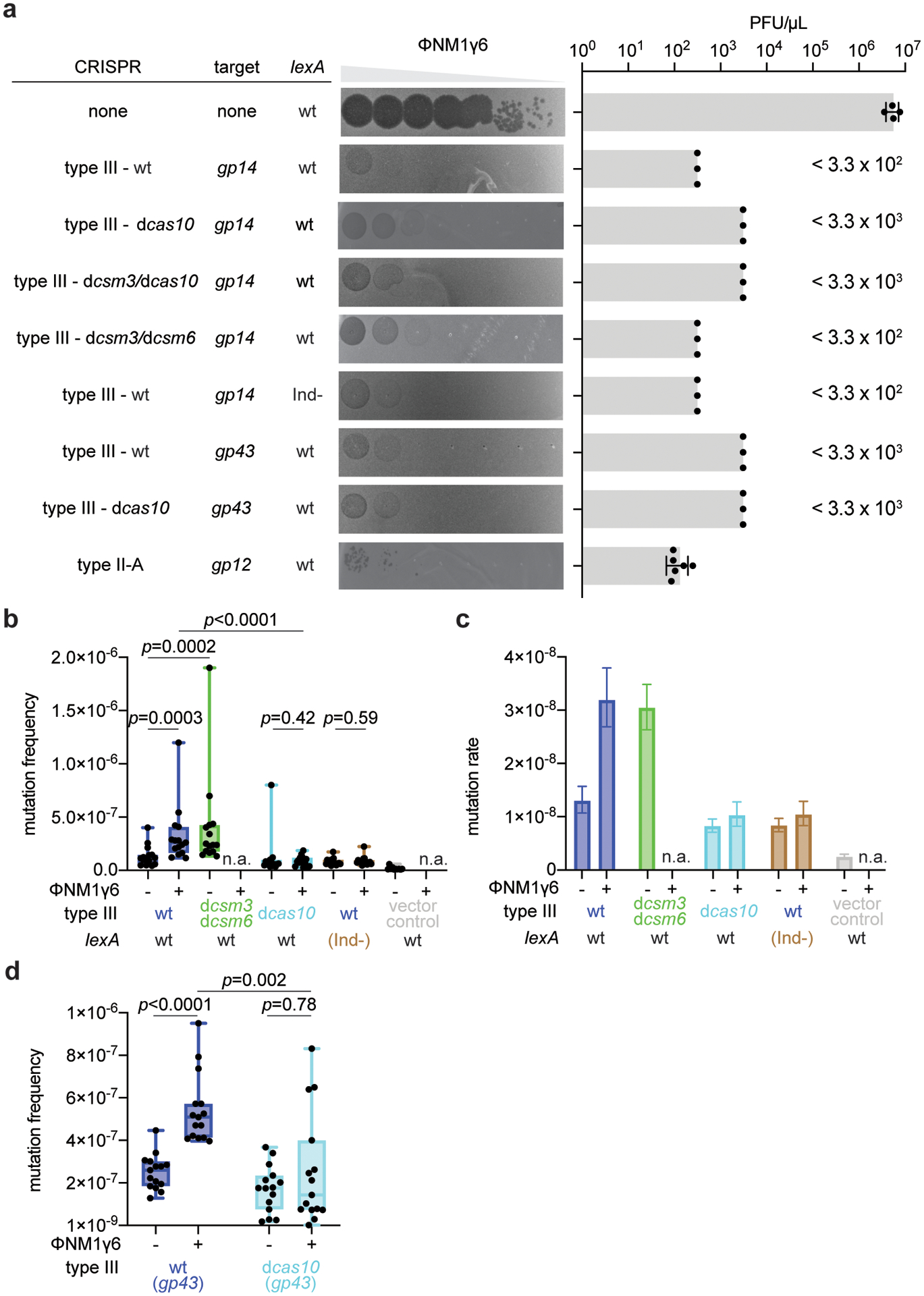
(a) Serial ten-fold dilutions of ϕNM1γ6 phage were spotted on plates seeded with different strains of staphylococci expressing a gp14- or gp43-targeting S. epidermidis type III-A systems with different mutations, wild-type or (Ind-) LexA, or the gp12-targeting S. aureus type II-A system. Plaque-forming units (PFU) were enumerated; mean ±s.e.m.; n=4 (non-targeting control) or n=6 (type II-A CRISPR immunity) biologically independent experiments. In cases where plaques were not readily visible, the limit of detection of three biologically independent experiments was reported. (b) S. aureus cells (<1,000) that were treated with ϕNM1γ6 phage and survived infection through the targeting of an early-expressed gene (gp14) by different mutant versions of the type III-A CRISPR-Cas system, in the presence of an active (wild-type, wt) or inactive (lexA(Ind-)) SOS response, were seeded on plates with or without rifampicin to calculate their mutation frequency. Box limits, interquartile range; whiskers, minimum to maximum; centre line, mesdian; dots, individual data points; n=15 biologically independent experiments; p-values obtained with two-sided Mann-Whitney test. (c) Calculation of the mutation rate using the data presented in (b). The bar graphs represent the mean; the error bars represent 95% confidence intervals. (d) S. aureus cells (~109) that were treated with ϕNM1γ6 phage and survived infection through the targeting of a late-expressed gene (gp43) by either a wild-type or a dcas10 type III-A CRISPR-Cas system, were seeded on plates with or without rifampicin to calculate their mutation frequency. Box limits, interquartile range; whiskers, minimum to maximum; centre line, median; dots, individual data points; n=15 biologically independent experiments; p-values obtained with two-sided Mann-Whitney test.
