Extended Data Fig. 5. Localizations of M9-βGal and NLS-2xBFP Complexes.
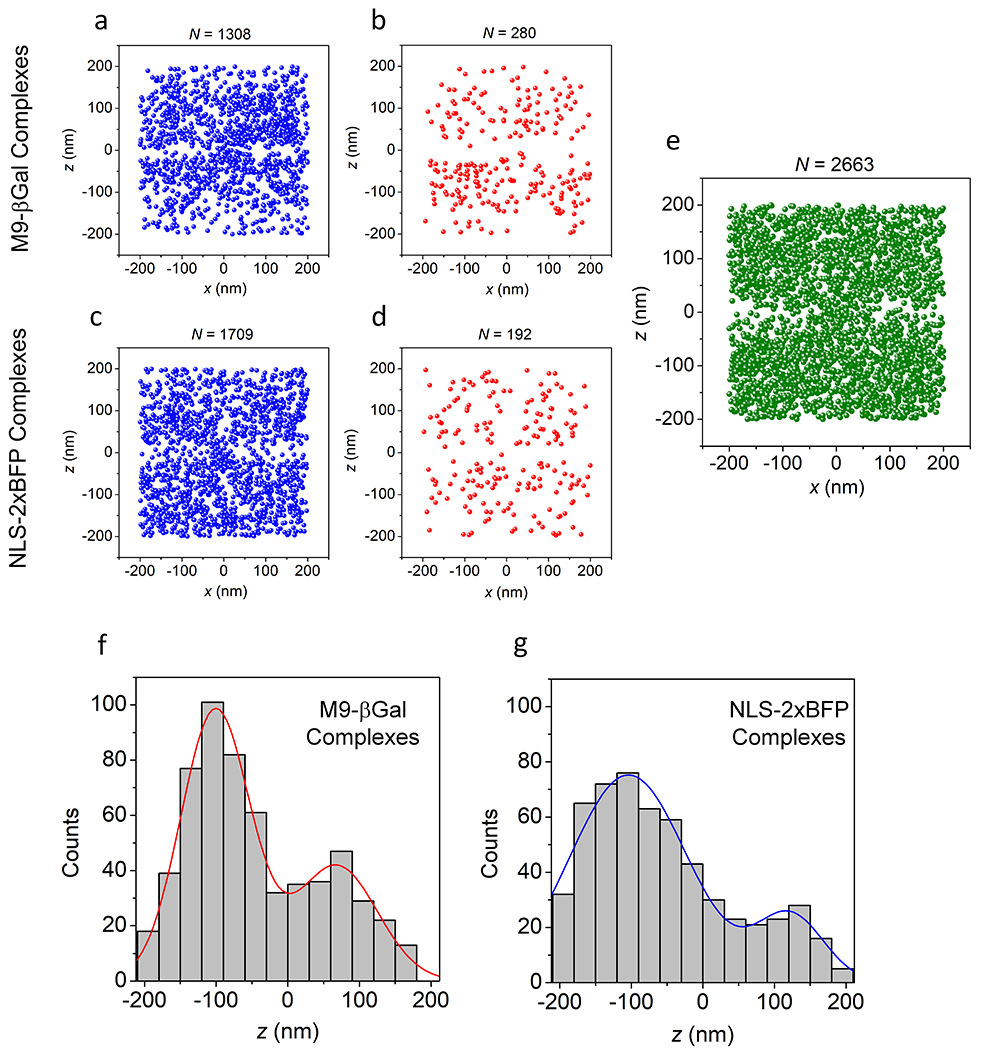
a,b, Brief M9-βGal complex appearances. Shown are the localizations from Fig. 4a that remained visible for one (a) or two (b) 3 ms frames. c,d, Brief NLS-2xBFP complex appearances. Shown are the localizations from the experiment in Fig. 5 that remained visible for one (c) or two (d) 2 ms frames. Localizations shown in (a-d) appear randomly distributed, consistent with particles that are largely diffusing and not interacting with the NPC. e, Simulated distribution of particles penetrating a barrier. In this simulation (Supplementary Data 2_Software File 2), particles appeared randomly within two compartments separated by a 50 nm thick barrier with a 100 nm diameter pore. This distribution models the M9-βGal complex localizations in Fig. 4a. The localizations appearing within the barrier result from the precision error, indicating that the localizations observed within the NE region in Fig. 4a are likely a consequence of localization error. For a-e, the N values are the number of localizations from 142 NPCs from 10 nuclei; each nucleus was an independent biological replicate. f,g, Localizations along the transport axis. Distribution of z values for M9-βGal complexes (f; from Figs. 4e,f) and NLS-2xBFP complexes (g; from Figs. 5e,f) undergoing import or abortive import. Distributions are fit to a double Gaussian function yielding mean values (±SD) of −100±48 nm and 68±56 nm for M9-βGal complexes, and −104±82 nm and 122±47 nm for NLS-2xBFP complexes. Source numerical data are provided in source data.
