Figure 5: BRD2 directly regulates transcription of interferon-induced genes.
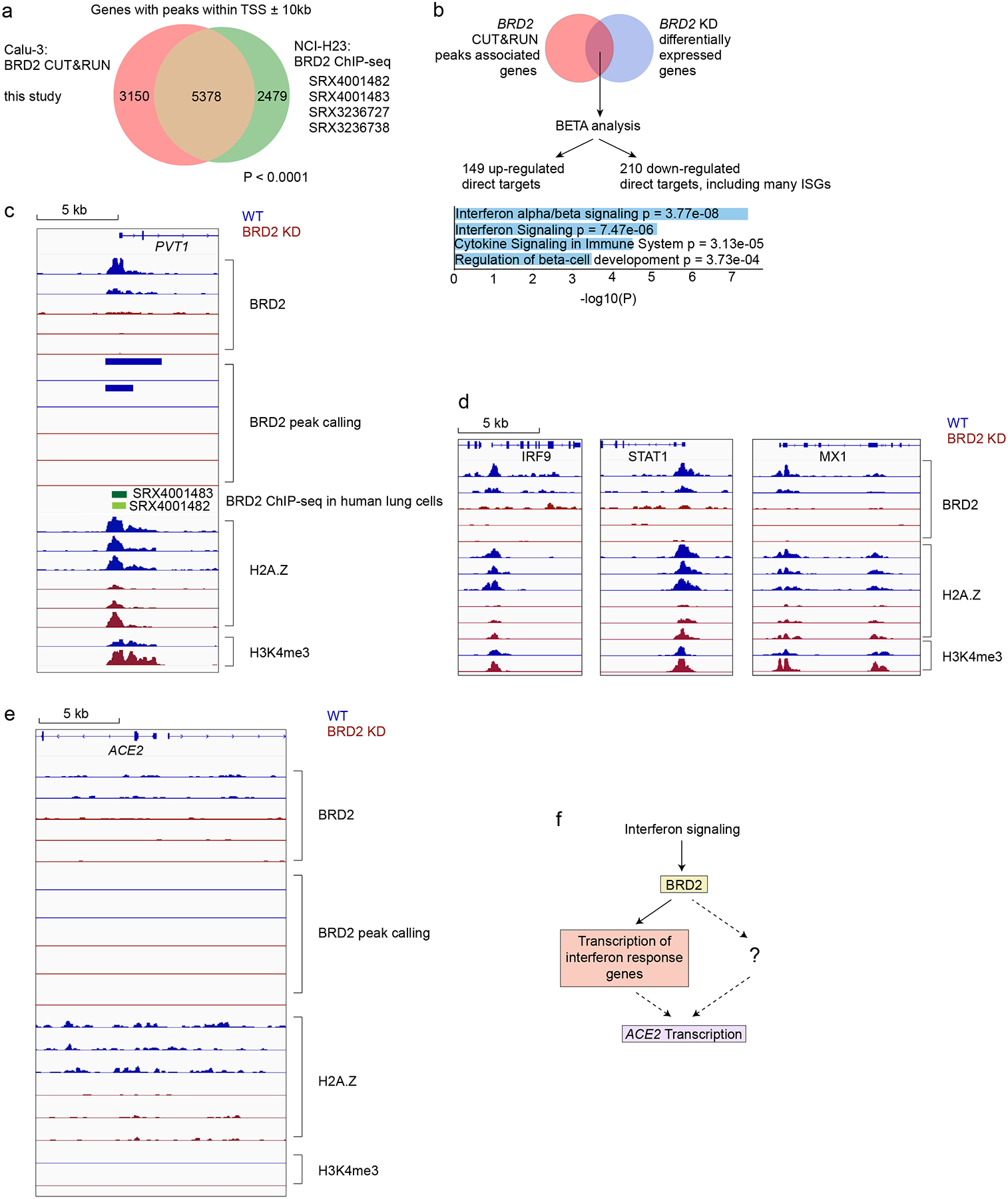
a, Genes associated with BRD2 CUT&RUN peaks within 10kb of a transcription start site determined in this study in Calu-3 cells overlap significantly with published BRD2 ChIP-seq peaks from the indicated datasets (P < 0.0001, two-sided Fisher’s exact test). b, Binding and Expression Target Analysis (BETA) was performed to identify direct BRD2 targets that were differentially expressed upon BRD2 knockdown using one-tailed Kolmogorov-Smirnov test. Many interferon response genes were identified as direct BRD2 targets. Direct BRD2 targets that were downregulated upon BRD2 knockdown were analyzed by ENRICHR for enriched Reactome pathways. Pathways with adjusted p-values less than 0.05 are displayed. c-e CUT&RUN experiments were conducted to map BRD2, H2A.Z and H3K4me3 genomic localization in WT (blue traces) and BRD2 KD (red) Calu-3 cells. Each trace represents an independent biological replicate. c, Known BRD2 regulatory sites are recapitulated. Raw signal tracks for WT and BRD2 knockdown cells are shown at the known BRD2 locus PVT1. BRD2 CHIP-seq tracks from human lung cells are shown. BRD2 peaks were called over IgG using SEACR at FDR < 0.05. d, Identified BRD2 and Histone H2A.Z occupancy and peak calling at ISGs in BRD2 KD and WT Calu-3 cells. Raw signal tracks for BRD2, Histone H2A.Z, and H3K4me are shown. BRD2 peaks were called over IgG using SEACR at FDR < 0.05. BRD2 CHIP-seq tracks from human lung cells are also shown. e, Raw BRD2 signal tracks and peak calling as for c, at the ACE2 locus. f, Proposed model for BRD2 control of ACE2 expression.
