FIG 1.
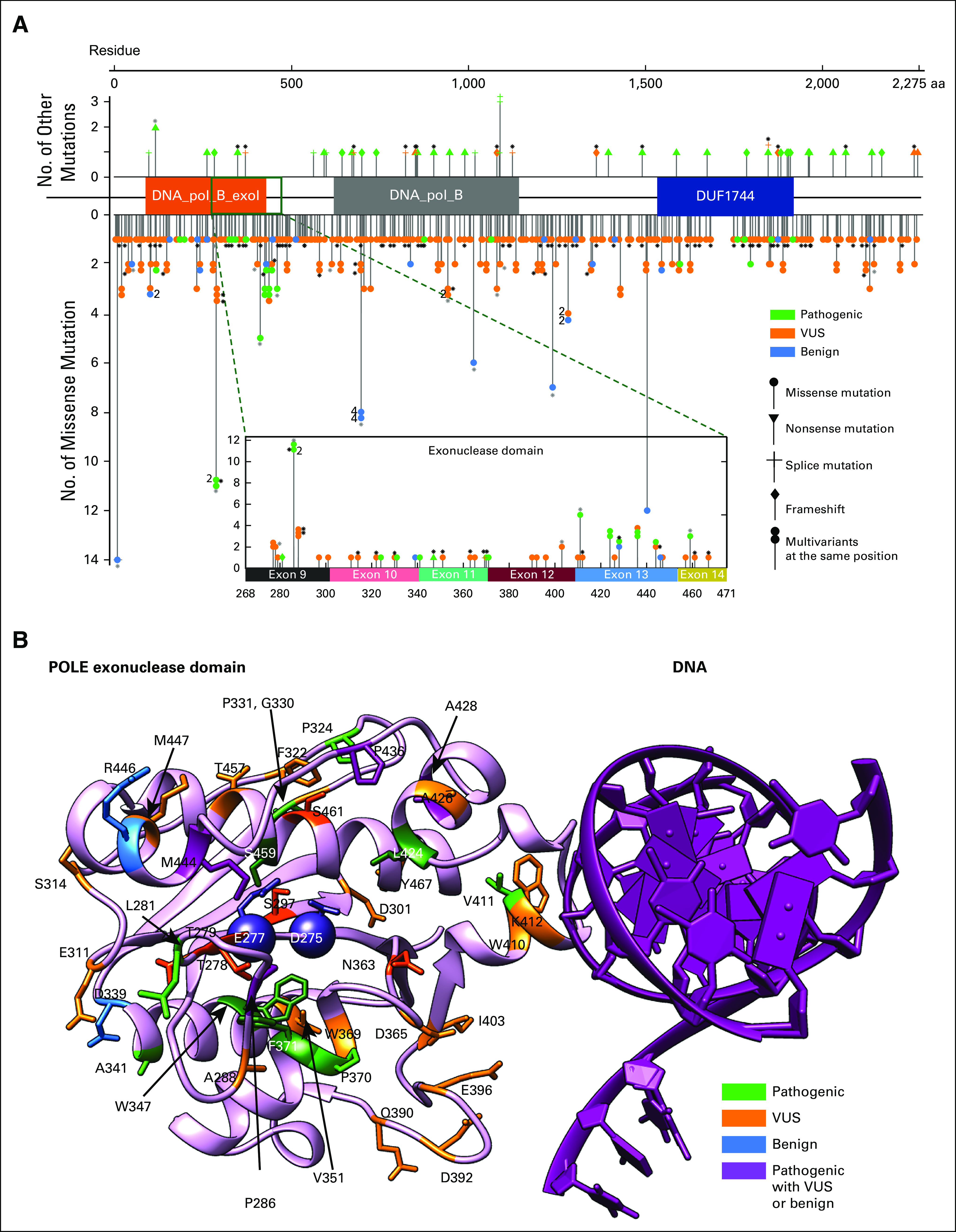
(A) Distribution of POLE mutations within POLE whole-length sequence. Among 450 evaluable patients, one had a POLE amplification and 449 had a POLE mutation, contributing to 424 unique POLE variants, as plotted against the mutational sites. Each data point with certain symbol represents a unique variant. All 375 unique missense mutations are shown downward with solid circles. Thirteen frameshift mutations, 13 splice variants, 23 nonsense mutations are plotted upwards in respective symbols. Stacked symbols indicate different mutations have been found at the same position. For example, POLE_P286 has 11 missense mutations occurrences among 449 patients in total, with one pathogenic mutation (P286L) in two patients and another VUS mutation found in nine patients. Black asterisks next to some data points indicate that those variants are in patients containing multiple POLE mutations, while gray asterisks represent a mixture of patients with a single POLE mutation and multiple POLE mutations. Fifty unique variants (47 missense mutations) are found within the exonuclease domain (268-471). (B) POLE exonuclease domain mutations mapped to structure. The exonuclease domain of human POLE modeled by AlphaFold229 (pink) is aligned to Saccharomyces cerevisiae POLE-DNA complex30 (DNA in purple) and then the POLE chain from Saccharomyces cerevisiae is removed. The two catalytic residues (D275 and E277) are shown as spheres. Mutations found in this domain are displayed in the structure with different colors indicating pathogenic status. Residues that are physically adjacent to the catalytic sites (all atom distance < 6 Å) are highlighted in darker colors, with VUS mutations S297, T278, T279, N363, and S461 in dark orange and pathogenic mutations M444, L424, S459, F371, W347, and P286 in dark green. These pathogenic mutations surround the two catalytic residues (D275 and E277)31 and likely affect the catalytic pocket, whereas pathogenic mutations at V411 might affect DNA binding, although its location is distal to the catalysis center. All benign mutations occur at residues far from catalytic sites. Other residues W410, I403, D365, D396, and D392 may also contribute to DNA binding, although the functional annotation of those mutations remains unknown. Different mutations at A428 (A428T and A428S) lead to conflicting pathogenic status in available databases; no patient treated with anti–PD-1/L1-based therapy in this cohort had an A428 mutation. PD-1, programmed death 1; PD-L1, programmed death ligand-1; VUS, variant of unknown significance.
