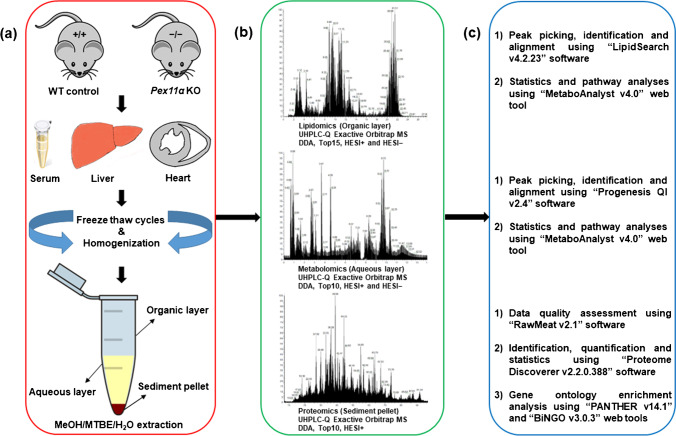
Fig. 1.
Schematic representation of the experimental workflow for sequential omics analyses of serum, liver, and heart tissue specimens from wild type (WT; Pex11α+/+) control and Pex11α knockout (KO; Pex11α−/−) mice. a Sample collection, homogenization, and simultaneous extraction of lipids (organic layer), polar metabolites (aqueous layer), and proteins (sediment pellet) using MeOH/MTBE/H2O liquid–liquid extraction. b Analytical workflow of multiple-molecular omics (lipidomics, metabolomics, and proteomics) data acquisition independently in positive- and negative-ion mode using ultra-high-performance liquid chromatography coupled with high-resolution tandem mass spectrometry (UHPLC-HRMS/MS). c Data processing, compound identification, statistics, and pathway analysis using dedicated bioinformatics tools