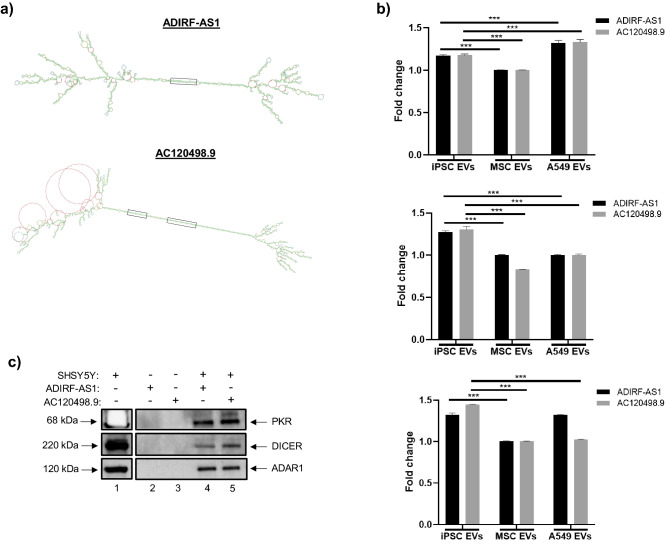
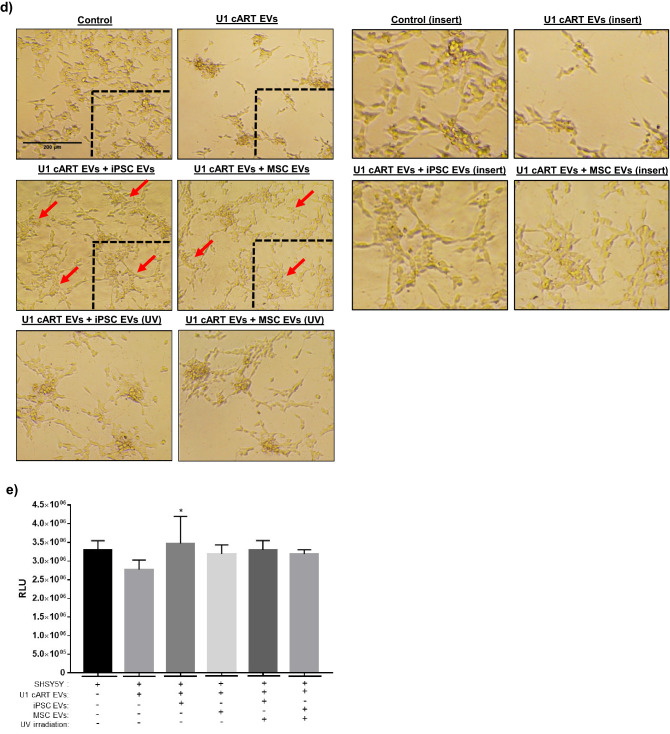
Figure 7.
RNA pulldown assay. (a) The secondary structures of lncRNAs ADIRF-AS1 and AC120498.9 are shown. These lncRNAs contained continuous stretches of dsRNA ≥ 30 base pairs in length; for reference, the corresponding regions are enclosed in black boxes. (b) Total RNA was isolated from EVs and RT-qPCR was carried out using primers that were specific to the boxed sequences shown in panel a. Data were normalized to GAPDH and the relative expression levels were evaluated using the 2−ΔΔCT method. n = 3. * p < 0.05, *** p < 0.0001. (c) Biotin-conjugated synthetic RNA sequences corresponding to the sequences of interest were incubated with SHSY5Ywhole cell extract. Western blot was performed to assess the expression of different RNA binding proteins (PKR, Dicer, ADAR1). (d) SHSY5Y cells were exposed to EVs from cART-treated, HIV-1 infected cells (U1) with or without treatment of stem cell EVs at an approximate ratio of 1:1000 (recipient cell to EV ratio). Stem cell EVs were exposed to UV(C) to inactivate EV-associated RNAs. Representative images show the appearance and morphology of cells after 8 days in culture. Inserts are included for higher resolution. Scale bar = 200 µm. n = 3. (e) Cell viability was quantified via CellTiter-Glo. n = 3. * p < 0.05 relative to U1 cART EVs.