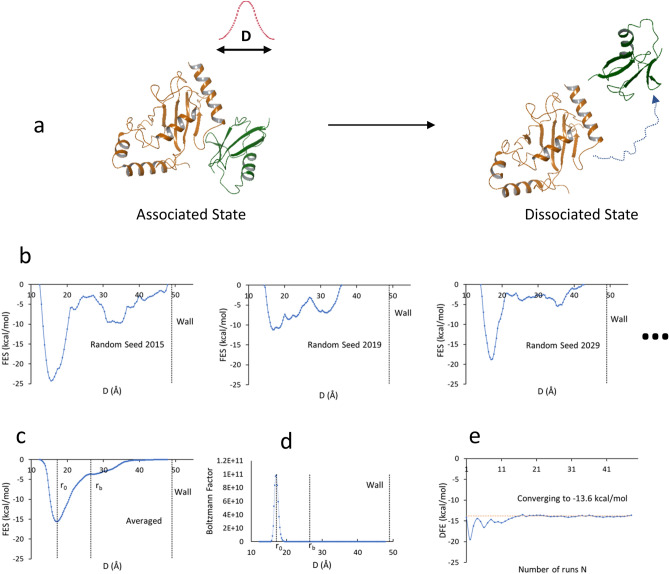
Figure 1.
Concept and workflow of a DFE calculation. (a) One-way trip dissociation of a molecular complex by running metadynamics using an intermolecular distance D as CV. The dotted curve on the right panel is an illustrative trajectory of the centroid of the CV residue of protein B (green ribbon). (b) Primitive FES from a number of replicas of different random seeds, noting that a wall is placed in each run. (c) Average FES stemming from an ensemble of primitive FES, noting the position r0 of the bound state minimum and the boundary rb separating the bound state region and the free state region. (d) Boltzmann factor as a function of D, narrowly concentrated around r0. (e) Convergence analysis: DFE is iteratively calculated as more and more replicas enter into averaging, showing that DFE converges when a sufficient number of replicas are averaged.