Fig. 6. TRα-dependent enrichment of cell cycle pathways among genes bound by TR in the wild-type intestine.
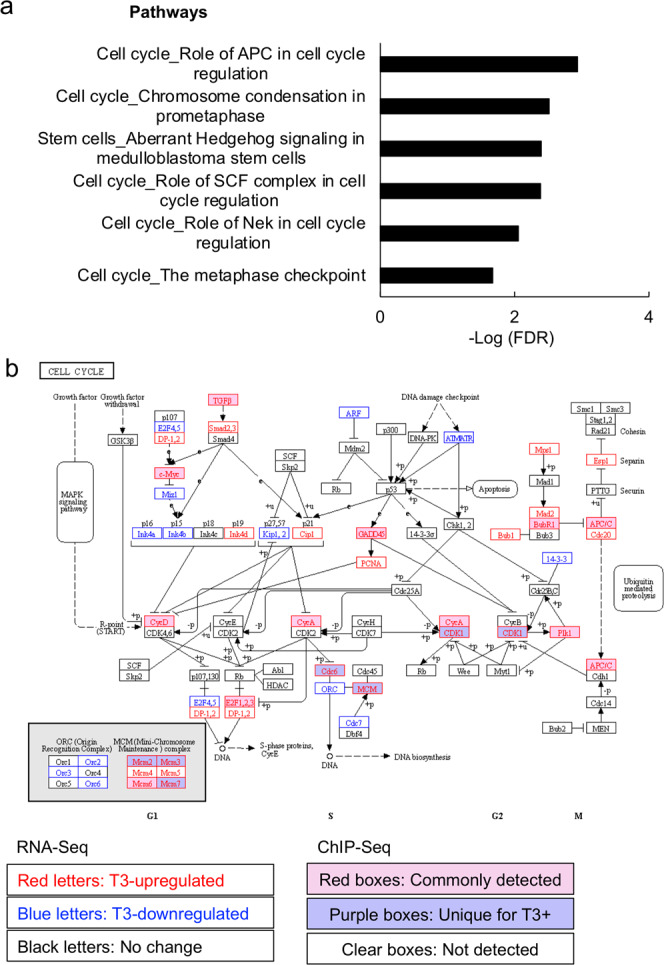
a Top 10 cell cycle pathways differentially enriched among genes bound by TR in wild-type intestine. The pathways that were enriched among genes bound by TR in the wild-type intestine (Supplementary Data 6) were subtracted by the pathways that were enriched among genes bound by TR in TRα−/− intestine (Supplementary Data 12) to obtain the pathways only enriched in the wild-type intestine (Supplementary Data 14). The most significant 10 cell cycle-related pathways were plotted here (see Supplementary Data 20 for raw data). b TR-binding and regulation of genes in the pathway for cell cycle based on ChIP-Seq and RNA-Seq data. The pathway for cell cycle was visualized with regard to genes regulated by T3 based on RNA-Seq and/or bound by TR based on ChIP-Seq. The arrows show functional interaction: green for activation. Letters in red and blue indicate upregulated and downregulated genes, respectively, in wild-type intestine after T3 treatment. Letters in black indicate genes with no change in wild-type intestine after T3 treatment. Red boxes indicate ChIP-Seq detected genes both with and without T3 treatment, and purple boxes indicate genes identified by ChIP-Seq only in wild type with T3 treatment. Note that Cyclin A and CDK1 were among the genes directly bound by TR and upregulated by T3.
