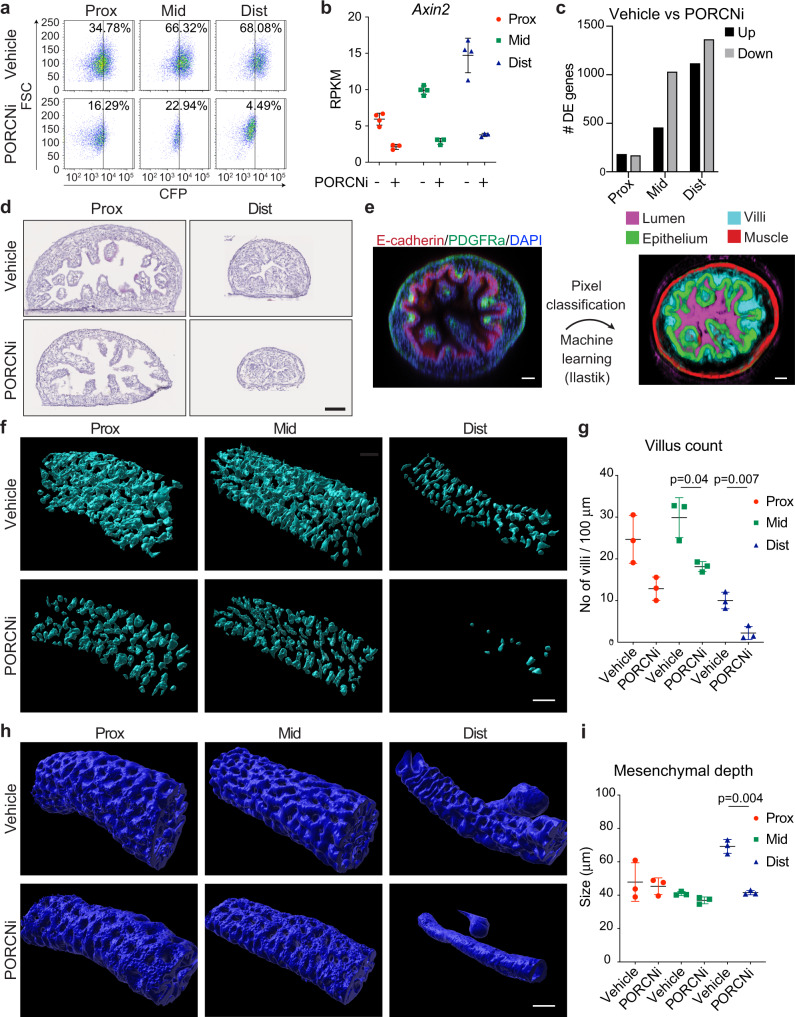
Fig. 3. Perturbation of Wnt-signalling has region-specific effects on SI transcription and structure.
a FACS profile showing the reduction of CFP+ cells (DAPI−/CD31−/CD45−/EpCAM+) in proximal, mid and distal regions upon PORCN inhibition (PORCNi). b RNA-seq data for Axin2 with and without PORCN inhibition. c RNA-seq analysis showing the number of differentially expressed genes comparing control and PORCNi samples for proximal, mid and distal regions. d H&E staining of proximal and distal regions in control and PORCNi samples. Scale bars, 100 μm. e Workflow for machine learning-assisted 3D image analysis. Scale bars, 40 μm. f Surface renderings of Ilastik-processed probability maps depicting villi in control and PORCNi samples. Scale bars, 100 μm. g Quantification of villus count in proximal, mid and distal regions in control and PORCN inhibited samples. h Surface renderings of Ilastik-processed probability maps depicting epithelium in control and PORCNi samples. Scale bars, 100 μm. i Quantification of mesenchymal depth in proximal, mid and distal regions in control and PORCNi samples. Results in a and d are representative of n = 3 independent experiments. Results in f and h are representative of n = 3 biological replicates. Data presented in b are means ± s.d. from n = 4 (Ctrl) or n = 3 (PORCNi) biological replicates. In g and i data presented are means ± s.d. from n = 3 biological replicates and statistical significance was assessed using two-tailed Welch’s t-test.