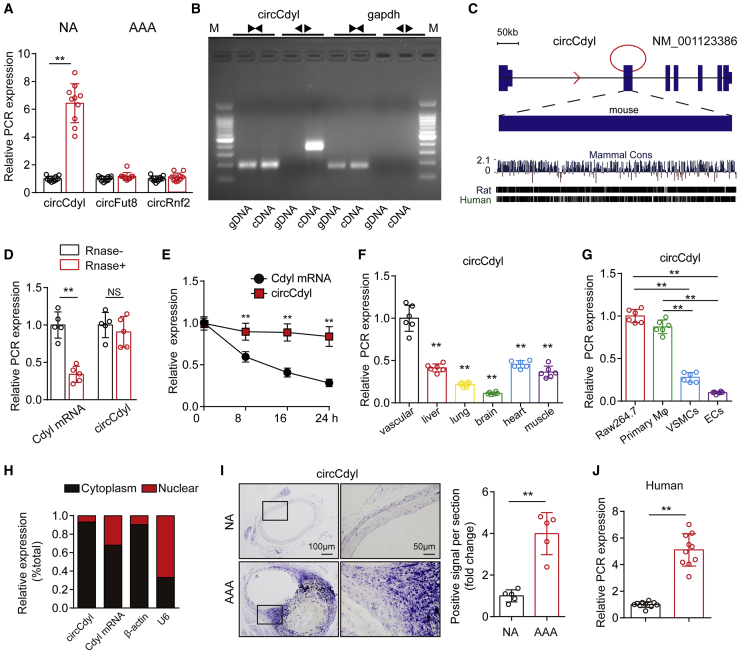
Figure 1.
circCdyl is significantly upregulated in AAA
(A) Relative expression of circCdyl, circFut8, and circRnf2 in mouse AAA and NA tissues (qPCR). ∗∗p < 0.01 versus the NA group; n = 10 per group. (B) Divergent primers amplify circCdyl from cDNA but not genomic DNA (gDNA); the divergent and convergent primers are indicated by the direction of the arrow. (C) Mouse genomic loci of the circCdyl in Cdyl genes. The expression of circCdyl was validated by reverse transcriptase polymerase chain reaction (RT-PCR) followed by Sanger sequencing. Its conserved analogs in humans, rats, and mice, as well as the per base conservation score, are also depicted. (D) Abundances of circCdyl and Cdyl mRNA in RAW264.7 cells treated with RNase R (RNase+) or untreated (RNase−). ∗∗p < 0.01 versus RNase−; NS, not significant versus RNase−; n = 5 per group. (E) Abundances of circCdyl and Cdyl mRNA in RAW264.7 cells treated with actinomycin D at the indicated time points (qPCR). ∗∗p < 0.01 versus Cdyl mRNA; n = 6 per group. (F) Relative expression of circCdyl in multiple mouse tissues (qPCR). ∗∗p < 0.01 versus vascular tissue; n = 6 per group. (G) Relative expression of circCdyl in several vascular cell types (qPCR). ∗∗p < 0.01 versus; n = 6 per group. (H) Abundance of circCdyl and Cdyl mRNA in either the cytoplasm or nucleus of RAW264.7 cells (qPCR). (I) In situ hybridization and densitometric analysis of aortic circCdyl in Ang II-treated mice (scale bars, 200 and 50 mm). ∗∗p < 0.01 versus NA; n = 5 per group. (J) Relative expression of circCdyl in human AAA and NA tissues (qPCR). ∗∗p < 0.01 versus NA; n = 10 per group.