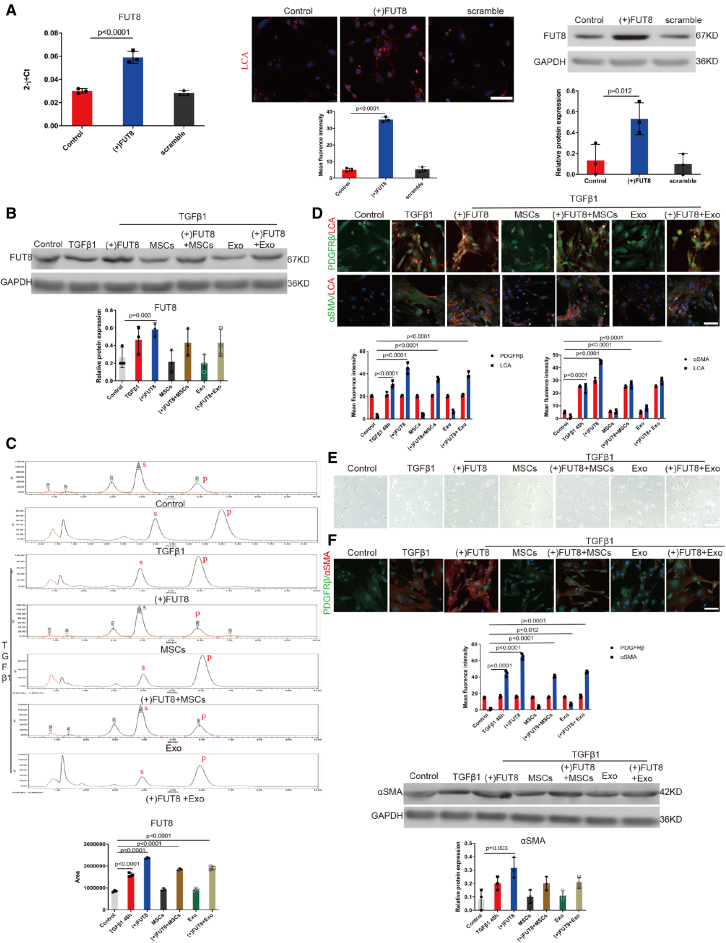
Figure 4.
MSC-derived exosomes inhibited pericyte activation mainly by regulating CF
(A) (Left) RT-PCR, (middle) representative images of LCA (red) staining, (right) FUT8 levels assessed by western blotting. The bottom panel shows the quantification. Data are the mean ± SD (n = 3). (B) FUT8 level was assessed by western blotting. The bottom panel shows the quantification. Scale bar, 50 μm. Data are the mean ± SD (n = 3). (C) FUT8 activity tested by HPLC. Data are the mean ± SD (n = 3); S indicates the peptide substrate; P is the fucosylation product. (D) Representative images of PDGFRβ (green) and LCA (red) staining, and α-SMA (green) and LCA (red) staining. The bottom panel shows the quantification. Scale bar, 75 μm. Data are the mean ± SD (n = 3). (E) Representative morphological alterations in pericytes. Scale bar, 200 μm. (F) Representative images of PDGFRβ (green) and α-SMA (red) staining. The bottom panel shows the quantification. Scale bar, 75 μm. Data are the mean ± SD (n = 3). α-SMA level was assessed by western blotting. The bottom panel shows the quantification. Scale bar, 50 μm. Data are the mean ± SD (n = 3).