Abstract
Pain sensitivity differs individually, but the mechanisms and genetic factors that underlie these differences are not fully understood. To investigate genetic factors that are involved in sensing cold pain, we applied a cold-induced pain test and evaluated protease-activated receptor 2 (PAR2/F2RL1) and transient receptor potential melastatin 8 (TRPM8), which are related to pain. We statistically investigated the associations between genetic polymorphisms and cold pain sensitivity in 461 healthy patients who were scheduled to undergo cosmetic orthognathic surgery for mandibular prognathism. We found an association between cold pain sensitivity and the rs2243057 polymorphism of the PAR2 gene. We also found a significant association between cold pain sensitivity and the rs12992084 polymorphism of the TRPM8 gene. Carriers of the minor A allele of the rs2243057 polymorphism of PAR2 and minor C allele of the rs12992084 polymorphism of TRPM8 exhibited a longer latency to pain perception in the cold-induced pain test, reflecting a decrease in cold pain sensitivity. These results suggest that genetic polymorphisms of both PAR2 and TRPM8 are involved in individual differences in cold pain sensitivity.
Keywords: PAR2, TRPM8, cold-induced pain test, cold pain sensitivity, single-nucleotide polymorphism
Introduction
Even with the same treatment, pain perception varies from person to person. Similar to sensitivity to analgesics, individual differences are observed in pain sensitivity. The influence of genetic factors on the sensitivity to fentanyl has been reported.1,2 However, the mechanisms and genetic factors that underlie differences in pain sensitivity are not fully understood.
Protease-activated receptor 2 (PAR2/F2RL1) is a seven-membrane-spanning, G-protein-coupled receptor.3,4 It is expressed in epithelial cells in many tissues throughout the body 3 and is known to be present in neurons of the human central nervous system (CNS), such as astrocytes, microglia, peripheral terminals of the central end of primary afferent nerve fibers, and posterior horn neurons.3,5–7
PAR2 is activated by trypsin, tryptase, blood coagulation factors (FVIIa, FXa), and neutrophil elastase3,4,8 through two pathways: (a) G protein and phospholipase C (PLC)/Ca2+/protein kinase C (PKC) signal transduction pathway and (b) β-arrestin-mediated extracellular signal-regulated kinase 1/2 (ERK1/2) activation pathway.3,5,9–13
Neural PAR2 is activated and regulates neural activity by being cleaved by mast cell-derived tryptase, which is in close contact with the choroid plexus, the parenchyma and perivascular regions of the CNS, and peripheral nerves. 3 PAR2 plays an important role in the response of organisms to tissue damage, especially in the process of inflammation and repair. 3 Many associations have been reported between PAR2 and pain, including mechanical allodynia and mechanical hyperalgesia, that is associated with cancer and irritable bowel syndrome in patients.3,5,9,13,14
The rs2243057 single-nucleotide polymorphism (SNP) of the PAR2 gene is a pain-related genetic variation. In a previous study, the rs2243057 SNP in the liver and whole blood was significantly associated with pleiotropic effects of dexamethasone, including a higher risk of osteonecrosis and thrombus, and an expression quantitative trait locus (eQTL) analysis suggested that this SNP affects changes in PAR2 mRNA levels.15,16 PAR2 is a pain-related molecule, and both osteonecrosis and thrombosis can cause pain as a primary symptom. The A allele of the rs2243057 of PAR2 was reported to be associated with an increase in PAR2 expression in the liver and whole blood. 15
The transient receptor potential (TRP) channel is an ion channel in the cell membrane that is involved in various biological functions as a sensor that senses temperature and chemical and physical stimuli. PAR2 activation has been reported to be involved in the sensitization of some TRP channels.3,5,17–19 Among TRP channels, transient receptor potential melastatin 8 (TRPM8) and transient receptor potential ankyrin 1 (TRPA1) are activated by cold stimulation, 20 although the functional association between PAR2 and these TRP channels has not yet been reported. TRPM8 and TRPA1 are activated at temperatures below 25 °C and 18 °C, respectively. TRPM8 belongs to the TRPM subfamily that consists of eight members. Three members of the TRPM subfamily (TRPM2, TRPM3, and TRPM8) are associated with pain. 21 TRPM8 is encoded by the TRPM8 gene and activated by low temperatures and many chemical agonists that are known to produce cold sensations, such as menthol, icillin, and eucalyptus.22–24 The functional association between PAR2 and TRPM8 has not yet been reported.
The present study used the cold-induced pain test to investigate genetic factors that are involved in sensing cold pain. We found a statistically significant association between the rs2243057 SNP of PAR2 and cold pain sensitivity. TRPM8 has the higher threshold for sensing cold temperatures relative to TRPA1. We also found that the rs12992084 SNP of TRPM8 was significantly associated with cold pain sensitivity. These results statistically suggested that both PAR2 and TRPM8 are involved in cold pain sensitivity. Together with the correlation between the genotype of the rs2243057 SNP of PAR2 and PAR2 expression level, we propose a molecular mechanism by which individual differences in cold pain sensitivity occur.
Materials and methods
Patients who were scheduled to undergo cosmetic orthognathic surgery
The protocol for this research project was approved by the Ethics Committees of Tokyo Dental College (approval no. 810) and conformed with the provisions of the Declaration of Helsinki. All of the subjects provided informed, written consent for the genetics studies. Enrolled in the study were 461 healthy patients (American Society of Anesthesiologists Physical Status I, age 15–58 years, 167 males and 294 females) who were scheduled to undergo cosmetic orthognathic surgery (mandibular sagittal split ramus osteotomy) for mandibular prognathism at Tokyo Dental College Suidobashi Hospital. All of the subjects were Japanese. Patients with chronic pain, who were taking pain medication, and who had experienced Raynaud’s phenomenon were excluded.
Preoperative cold-induced pain test
The patients were premedicated with oral diazepam (5 mg) and oral famotidine (50 mg) 90 min before the induction of anesthesia. The patients had an intravenous (i.v.) line inserted in the forearm on their nondominant side. The temperature in the operating room was maintained at 26 °C. The cold-induced pain test was then performed before and 3 min after an i.v. bolus injection of fentanyl (2 μg/kg) as previously described.25,26 Crushed ice cubes and cold water were blended 15 min before the test in a 5-L isolated tank, and the mixture was stirred immediately before each test to ensure uniform temperature distribution (0 °C) within the tank. The patients were instructed to immerse their dominant hand in the ice-cold water to the wrist, keep their hand calm, and immediately remove it when they perceived any pain. All of the patients were tested by the same investigator. The baseline latency to pain perception, defined as the time of immersion of the hand in the ice water before the i.v. injection of fentanyl (PPLpre), was recorded. A cut-off time of 150 s was set to avoid tissue damage. The hand was warmed with a hair dryer as soon as it was withdrawn from the ice water until the sensation of cold was completely abolished. The patients then received an i.v. injection fentanyl (2 μg/kg). Three minutes after the injection, the latency of pain perception of the dominant hand (PPLpost) was measured again. The difference between PPLpre and PPLpost (PPLpost–PPLpre) was defined as the preoperative analgesic effect.
Anesthesia and surgery
After the cold-induced pain test, general anesthesia was induced with a target-controlled infusion (TCI) of propofol using a TCI pump (TE-371, Terumo Corporation, Tokyo, Japan). Vecuronium (0.1 mg/kg) was administered to facilitate nasotracheal intubation. For the preparation of DNA specimens, 10 ml of venous blood was sampled after the induction of anesthesia. General anesthesia was maintained with propofol at a target blood concentration of 4–6 μg/ml, and vecuronium was administered at a rate of 0.08 mg/kg/h. The lungs were ventilated with oxygen-enriched air. Local anesthesia was performed on the right side of the surgical field with 8 ml of 2% lidocaine that contained 12.5 μg/ml epinephrine, and right mandibular ramus osteotomy was performed. Local anesthetic block and mandibular ramus osteotomy were then similarly performed on the left side, and the bilateral mandibular bone segments were fixed in appropriate positions. Whenever systolic blood pressure or heart rate exceeded +20% of the preinduction value during surgery, i.v. fentanyl, 1 μg/kg, was administered.
Postoperative pain management
At the end of surgery, rectal diclofenac sodium (50 mg) and i.v. dexamethasone (8 mg) were administered at the request of surgeons to prevent postoperative orofacial edema/swelling. After emergence from anesthesia and tracheal extubation, droperidol (1.25 mg) was administered i.v. to prevent nausea/vomiting. A fentanyl-droperidol combination (2 mg fentanyl and 5 mg droperidol diluted in normal saline in a total volume of 50 ml) was administered using a CADD-Legacy PCA pump (Smiths Medical Japan, Tokyo, Japan) for i.v. patient-controlled analgesia (PCA). The bolus dose of fentanyl on demand and lockout time were set at 20 μg and 10 min, respectively. Continuous background infusion was not employed. Droperidol was coadministered with fentanyl to prevent nausea/vomiting because our preliminary study showed that young females had a high incidence (up to 30%) of nausea/vomiting with PCA fentanyl. Patient-controlled analgesia was continued for 24 h postoperatively. In the case of treatment-refractory adverse effects or inadequate analgesia, PCA was discontinued, and rectal diclofenac sodium (50 mg) was prescribed as a rescue analgesic as required. A 100-mm visual analog scale (VAS), with 0 mm indicating no pain and 100 mm indicating the worst pain imaginable, was used to assess the intensity of spontaneous pain at 3 and 24 h postoperatively. Intraoperative fentanyl use, postoperative PCA fentanyl use during the first 24-h postoperative period, and perioperative (i.e., intraoperative + postoperative) fentanyl use were recorded, and the perioperative doses of fentanyl were normalized to body weight.
Genotyping
This study examined SNPs of the PAR2 and TRPM8 genes. The rs2243057 SNP of PAR2 was selected because associations between the rs2243057 of PAR2 and risk of osteonecrosis and thrombosis were previously reported. 15 For the SNPs of TRPM8, we analyzed 47 SNPs around the TRPM8 gene region (including 10 kbp upstream and downstream) using genotype data from whole-genome genotyping in 361 healthy patients who were scheduled to undergo cosmetic orthognathic surgery. 27 Whole-genome genotyping was performed using Infinium assay II and the iScan system (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Five kinds of BeadChips were used to genotype 40, 67, 6, 119, and 2 samples, respectively: HumanHap300 (total markers: 3,17,503), HumanHap300-Duo (total markers: 3,18,237), Human610-Quad v1 (total markers: 6,20,901), Human1M v1.0 (total markers: 10,72,820), and Human 1 M-Duo v3 (total markers: 11,99,187). Some BeadChips included a number of probes that were specific to copy number variation markers, but most were for SNP markers on the human autosome or sex chromosome. Approximately 3,00,000 SNP markers were commonly included in all of the BeadChips.
Genomic DNA was extracted from whole-blood samples using standard procedures. The extracted DNA was dissolved in TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8.0). The DNA concentration was adjusted to 5–50 ng/µl for genotyping the rs2243057 and rs12992084 SNPs by the TaqMan assay or 100 ng/µl for whole-genome genotyping using a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific K.K., Tokyo, Japan).
The TaqMan assay was performed on 461 samples of the rs2243057 SNP and on 100 samples of the rs12992084 SNP. To perform the TaqMan assay with a LightCycler 480 (Roche Diagnostics K.K., Tokyo, Japan), TaqMan SNP Genotyping Assays (Thermo Fisher Scientific K.K) were used that included sequence-specific forward and reverse primers to amplify the polymorphic sequence and two probes that were labeled with VIC and FAM dye to detect both alleles of the PAR2 and TRPM8 SNPs. The sequences of the primers for rs2243057 and rs12992084 were not disclosed. Real-time polymerase chain reaction (PCR) was performed in a final volume of 10 μl that contained 2 × LightCycler 480 Probes Master (Roche Diagnostics K.K.), 40 × TaqMan SNP Genotyping Assays, 5–50 ng genomic DNA as the template, and up to 10 μl H2O equipped with 2 × LightCycler 480 Probes Master. The thermal conditions were the following: 95 °C for 10 min, followed by 45 cycles of 95 °C for 10 s and 60 °C for 60 s, with final cooling at 50 °C for 30 s. Afterward, endpoint fluorescence was measured for each sample well, and the G/G, A/G, and A/A genotypes of rs2243057 and T/T, T/C, and C/C genotypes of rs12992084 were determined based on the presence or absence of each type of fluorescence.
Statistical analysis
The patients’ demographic and clinical data are expressed as mean ± SD. As reported in our previous study, the data had no bias in the distribution and were thus suitable as a group for the statistical analysis. 28 The statistical analysis was performed using SPSS 25 software (IBM Japan, Tokyo, Japan). For PAR2, the Kruskal-Wallis test and Mann-Whitney U-test were performed to detect possible associations between any of the genomic parameters and clinical endpoints related to pain sensitivity (i.e., PPLpre and PPLpost–PPLpre) or the analgesic effects of fentanyl (i.e., postoperative fentanyl use during 24 h [μg/kg] and VAS pain score at 3 and 24 h [mm]). The Jonckheere-Terpstra trend test was performed to investigate linear trends. In the statistical tests for the PAR2 SNP, the criterion for significance was set at p < 0.05. Genotype data that were extracted from the whole-genome genotyping for TRPM8 contained 47 SNPs. Therefore, Bonferroni correction for multiple comparisons was applied to p values, and significance was set at p < 0.0011 for TRPM8 SNPs.
We investigated the relationship between PPLpre, PPLpost–PPLpre, 24-h postoperative fentanyl use, and VAS pain scores at 3 and 24 h and the rs2243057 SNP of PAR2 gene. Forty-seven SNPs of TRPM8, which is a TRP channel-related gene, were extracted from the genotyping data of whole-genome genotyping of 361 samples. The significantly associated polymorphisms were further analyzed together with an additional 100 samples that were genotyped using the TaqMan assay (Mann-Whitney U-test and Spearman’s rank correlation test). Bonferroni correction for multiple comparisons was applied for the SNPs of TRPM8.
Results
Cold water-induced pain sensitivity was associated with rs2243057 SNP of PAR2 gene
Although PAR2 is associated with nociceptive pain, cancer pain, and neuropathic pain,3,5,8 the association between cold pain and PAR2 has not yet been reported. To clarify the association between cold pain and PAR2, we investigated the relationship between the rs2243057 SNP of PAR2 and PPLpre, PPLpost–PPLpre, 24-h postoperative fentanyl use, and VAS pain scores at 3 and 24 h. All of the 461 Japanese patients with postoperative pain who were enrolled in the study completed the study. The patients’ demographic and clinical data are shown in Table S1. The patients’ genotype distributions of the SNP are shown in Table 1.
Table 1.
Genotype distribution of SNPs for patients who underwent cosmetic orthognathic surgery.
| Gene | SNP | Genotype | ||
|---|---|---|---|---|
| PAR2 | rs2243057 | GG : 247 (53.6%) | AG : 185 (40.1%) | AA : 29 (6.3%) |
| TRPM8 | rs12992084 | TT : 389 (84.4%) | TC : 70 (15.2%) | CC : 2 (0.4%) |
Data are expressed as the number (%) of subjects.
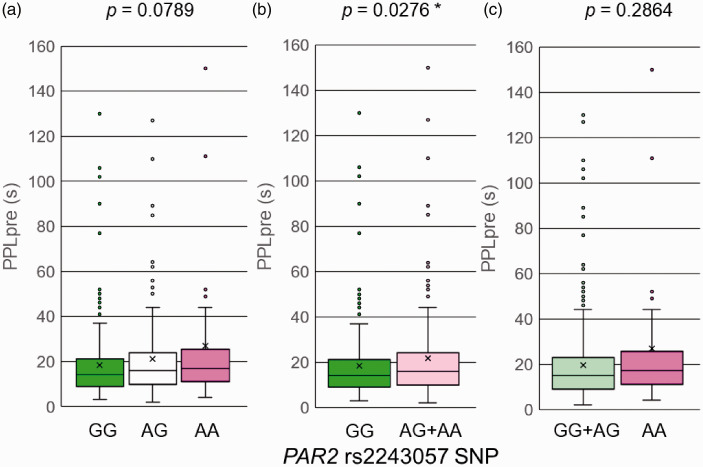
For PPLpre, a significant difference was found between the GG group and AG + AA group (p = 0.0276; Mann-Whitney U-test; Table 2; Figure 1). The mean values of PPLpre in the GG group and AG + AA group were 18.2 and 21.8 s, respectively. The AG + AA group had a longer latency to pain perception before the i.v. injection of fentanyl (PPLpre) compared with the GG group. PPLpre indicated baseline sensitivity to cold pain perception. These results raise the possibility that the GG genotype of the rs2243057 SNP is associated with higher sensitivity (i.e., a lower threshold) to cold pain. When we divided patients into GG, AG, and AA groups, a trend toward an association with PPLpre values was found among each genotype group (GG, AG, and AA, p = 0.0789; Kruskal-Wallis test; Table 2; Figure 1). To clearly demonstrate linearity of the cold pain threshold by copy number of the A allele of the SNP, we applied the Jonckheere-Terpstra trend test, which revealed a positive correlation between PPLpre values and copy number of the A allele of the SNP (p = 0.0238; Jonckheere-Terpstra trend test; Table 2). Thus, the threshold of cold pain linearly increased as the copy number of the minor A allele of the SNP increased. When we divided the patients into the GG + AG group and AA group, we did not find a significant difference in PPLpre between groups (p > 0.05; Mann-Whitney U-test; Table 2; Figure 1). For PPLpost–PPLpre, 24-h postoperative fentanyl use, and VAS pain scores at 3 and 24 h, no significant differences were observed among each genotype group (GG, AG, and AA, p > 0.05; Kruskal-Wallis test; Table 2), between the GG group and AG + AA group, or between the GG + AG group and AA group for the rs2243057 SNP (p > 0.05; Mann-Whitney U-test; Table 2).
Table 2.
Association analysis between clinical data and PAR2 gene rs2243057 SNP (p value).
| GG/AG/AA | GG/AG/AA | GG/AG + AA | GG + AG/AA | |
|---|---|---|---|---|
| Phenotype | (Kruskal-Wallis test) | (Jonckheere-Terpstra trend test) | (Mann-Whitney U test) | (Mann-Whitney U test) |
| PPLpre (s) | 0.0789 | 0.0238* | 0.0276* | 0.2864 |
| PPLpost (s) | 0.2317 | 0.0948 | 0.1186 | 0.2858 |
| PPLpost-PPLpre (s) | 0.6183 | 0.3893 | 0.4616 | 0.4083 |
| 24-h postoperative fentanyl use (μg/kg) | 0.7617 | N/A | 0.7348 | 0.5964 |
| VAS pain score at 3 h (mm) | 0.3484 | N/A | 0.2737 | 0.2188 |
| VAS pain score at 24 h (mm) | 0.9139 | N/A | 0.7192 | 0.7491 |
VAS: visual analog scale; PPL: latency to pain perception; N/A: not applicable. *p < 0.05.
Figure 1.
Associations between genotypes of the rs2243057 SNP of PAR2 and latency to pain perception before fentanyl administration (PPLpre) in the cold-induced pain test. The data are expressed as box and whisker plots. The upper and lower ends of the boxes represent the 75th and 25th percentiles, respectively. Whiskers represent the highest and lowest values. The medians are depicted by horizontal solid lines in the boxes. The mean values are depicted by crosses. Outliers are shown as circles. *p < 0.05. (a) The samples were divided into three groups (GG, AG, and AA). (b) The samples were divided into two groups (GG and AG + AA). (c) The samples were divided into two groups (GG + AG and AA).
Cold water-induced pain sensitivity was associated with rs12992084 SNP of TRPM8 gene
To determine whether TRPM8 is related to cold pain sensitivity, we investigated associations between the SNPs of TRPM8 and cold pain sensitivity using clinical samples.
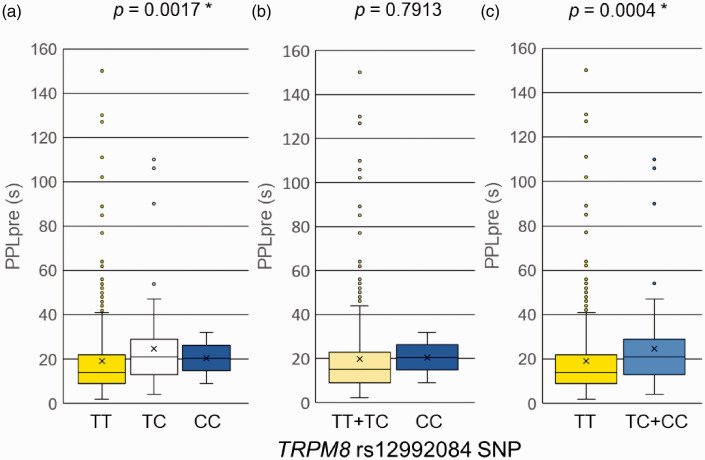
To explore the SNP of TRPM8 that had a significant association with PPLpre, a portion of genotype data for TRPM8 polymorphisms was undiscriminatingly extracted from the entire whole-genome genotyping data that were obtained in previous genome-wide association studies (GWASs)1,27,29 because no previous studies have reported polymorphisms of the TRPM8 gene. Genotype data for a total of 47 SNPs, located in and around the TRPM8 gene, were extracted from the entire whole-genome genotyping data from 361 healthy patients in the association study with PPLpre (i.e., the phenotype that was associated with the rs2243057 SNP of PAR2). We did not further analyze associations between SNPs of TRPM8 and PPLpost–PPLpre, 24-h postoperative fentanyl use, or VAS pain scores at 3 and 24 h because of the lack of significant associations between these phenotypes and the rs2243057 SNP of PAR2. Among the 47 SNPs of the TRPM8 gene, rs12992084 was significantly associated with PPLpre (p = 0.0009; the level of significance was set at p < 0.0011 after Bonferroni correction for multiple comparisons thereafter). Because the rs12992084 SNP had a significant association with PPLpre in 361 samples in the initial analysis, a further statistical analysis was performed with a larger sample size for this SNP. As a result of genotyping and the statistical analysis of TRPM8 in 461 samples, a significant difference was found between the TT group and TC + CC group in PPLpre (p = 0.0004; Mann-Whitney U-test; Table 3; Figure 2). When we divided the patients into the TT + TC group and CC group, we did not find a significant difference in PPLpre between groups (p > 0.0011; Mann-Whitney U-test; Table 3; Figure 2). When we divided the patients into the TT, TC, and CC groups, a trend toward an association with PPLpre was found among each genotype group (TT, TC, and CC, p = 0.0017, 0.0011 < p < 0.0021 was considered a trend toward an association after Bonferroni correction for multiple comparisons; Kruskal-Wallis test; Table 3; Figure 2). To ascertain linearity of the cold pain threshold by copy number of the C allele of the SNP, we applied the Jonckheere-Terpstra trend test, which revealed a positive correlation between PPLpre and copy number of the C allele of the SNP (p = 0.0004; Jonckheere-Terpstra trend test; Table 3). Thus, the threshold of cold pain linearly increased as the copy number of the minor C allele of the SNP increased. These results suggest that the CC genotype of the rs12992084 SNP is associated with lower sensitivity to cold pain.
Table 3.
Association analysis between PPLpre and TRPM8 gene rs12992084 SNP (p value).
| TT/TC/CC | TT/TC/CC | TT + TC/CC | TT/TC + CC | |
|---|---|---|---|---|
| Phenotype | (Kruskal-Wallis test) | (Jonckheere-Terpstra trend test) | (Mann-Whitney U test) | (Mann-Whitney U test) |
| PPLpre (s) | 0.0017 | 0.0004* | 0.7913 | 0.0004* |
PPL: latency to pain perception. *p < 0.0011 after Bonferroni correction for multiple comparisons.
Figure 2.
Associations between genotypes of the rs12992084 SNP of TRMP8 and the latency to pain perception before fentanyl administration (PPLpre) in the cold-induced pain test. The data are expressed as box and whisker plots. The upper and lower ends of the boxes represent the 75th and 25th percentiles, respectively. Whiskers represent the highest and lowest values. The medians are depicted by horizontal solid lines in the boxes. The mean values are depicted by crosses. Outliers are shown as circles. *p < 0.0011 after Bonferroni correction for multiple comparisons. (a) The samples were divided into three groups (TT, TC, and CC). (b) The samples were divided into two groups (TT + TC and CC). (c) The samples were divided into two groups (TT and TC + CC).
Discussion
PAR2 and TRP channels have been reported to be associated with neuropathic pain and hyperalgesia.3,5,13,14,21 PAR2 is associated with transient receptor potential vanilloid 1 (TRPV1) and TRPV4, but the relationship between PAR2 and TRPM8 has not yet been reported. In the present study, we found that genetic polymorphisms of the PAR2 and TRPM8 genes were associated with cold pain sensitivity. Carriers of the minor A allele of the rs2243057 SNP of PAR2 and minor C allele of the rs12992084 SNP of TRPM8 had a longer latency to pain perception, indicating lower cold pain sensitivity. The results showed that carriers of minor alleles of SNPs of both PAR2 and TRPM8 had lower sensitivity to cold pain. However, we could not clarify the mechanisms by which this occurred because we only analyzed gene polymorphisms. Further studies are also needed that increase the number of subjects to obtain more reliable data.
Although the relationship between PAR2 and TRPV1/TRPV4 has been reported,3,5,17–19 these TRPVs are unlikely to react to cold stimulation because of the high threshold of temperature sensing. Our experiment was conducted using cold stimulation. Thus, TRPM8 and TRPA1 may participate in cold sensing as cold receptors. Based on the earlier reaction to cold stimulation, we focused on TRPM8 and found a significant association between the SNP of TRPM8 and cold pain sensitivity. Several signal transduction pathways that involve TRPM8 have been reported, including the nerve growth factor-TrkA signal transduction pathway30,31 and PKC pathway, mediated by phosphatidylinositol 4,5-bisphosphate (PIP2).24,30,32,33 In PAR2-related signal transduction pathways, PIP2 is depleted in accordance with PAR2 activation 34 in G protein and PLC/Ca2+/PKC signaling pathways.3,5,9–13 Accordingly, TRPM8 and PAR2 are likely to be related via PIP2 in the PLC/Ca2+/PKC signaling pathway. Moreover, the PLC/Ca2+/PKC signaling pathway likely plays a major role in cold pain sensing. TRPM8 channel activity may be inhibited by a decrease in PIP2 levels through TRPM8 activation, PIP2 hydrolysis by PLC, PKC activation, and an increase in intracellular Ca2+ levels. Additionally, PAR2 activation may also decrease PIP2 levels and inhibit TRPM8 channel activity. As a result, the transmission of cold pain via TRPM8 may be inhibited by PAR2 activation, thereby lowering cold pain sensitivity (Figure S1). However, further direct evidence is needed to confirm this hypothesis.
A previous study suggested that carriers of the AA genotype of the rs2243057 of PAR2 have higher PAR2 expression levels. 15 Carriers of the AA genotype of the rs2243057 SNP of PAR2 had lower sensitivity to cold-induced pain. The expression level of PAR2 is higher in A allele carriers than in G allele carriers. The inhibitory effects of TRPM8 activity are stronger, and sensitivity decreases because of the higher depletion of PIP2 by cold stimulation in A allele carriers than in G allele carriers (Figure S1).
The rs12992084 SNP is located in the 7th intron region of the TRPM8 gene on chromosome 2p12. This SNP in the intron region may affect the transcriptional activity of TRPM8, perhaps resulting in alterations of TRPM8 protein expression levels. Lower sensitivity was observed in carriers of the C allele of the rs12992084 SNP of TRPM8 in the cold-induced pain test. TRPM8 protein expression levels may be lower in C allele carriers than in noncarriers, based on the logic mentioned above (Figure S1). However, the relationship between this SNP and TRPM8 expression levels needs to be clarified.
Although we analyzed the relationship between PPLpre and 32 SNPs of TRPA1 (i.e., another cold receptor gene), the data for which were extracted from whole-genome genotyping data, we did not find a significant association (data not shown). del Camino et al. suggested that TRPA1 is a key mediator of cold hypersensitivity under pathological conditions but likely plays a comparatively minor role in acute cold sensation. 35 TRPA1 may begin to react with prolonged cold exposure because TRPM8 and TRPA1 are not co-expressed in the same nerve cells, and TRPA1 is activated by a lower temperature than TRPM8. 36
In conclusion, the present study found that cold pain sensitivity was associated with SNPs of PAR2 and TRPM8. Together with previous observations, the present findings suggest that PAR2 and TRPM8 are involved in cold pain sensing via common signaling molecules in both the PAR2 and TRPM8 signal transduction pathways. Direct evidence of this relationship needs to be elucidated in future studies.
Supplemental Material
Supplemental material, sj-pdf-1-mpx-10.1177_17448069211002009 for Cold pain sensitivity is associated with single-nucleotide polymorphisms of PAR2/F2RL1 and TRPM8 by Moe Soeda, Seii Ohka, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Tatsuya Ichinohe, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental material, sj-xlsx-2-mpx-10.1177_17448069211002009 for Cold pain sensitivity is associated with single-nucleotide polymorphisms of PAR2/F2RL1 and TRPM8 by Moe Soeda, Seii Ohka, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Tatsuya Ichinohe, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Acknowledgements
We thank Mr. Michael Arends for his assistance with editing the manuscript. We are grateful to the volunteers for their participation in the study and anesthesiologists and surgeons for collecting the clinical data.
Footnotes
Authors’ Contributions: MS, SO, DN, and KI conceived the study and designed the experiments. MS, SO, KN, YE, and DN performed the statistical analyses. MS wrote the manuscript. KF and TI collected clinical samples and data. MS and JH performed the genotyping procedures. SO, DN, and KI supervised the experiments and finalized the manuscript. All of the authors contributed to writing the manuscript, and all of the authors read and approved the final manuscript.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the Japan Society for the Promotion of Science (JSPS) KAKENHI (no. JP16H06276 [AdAMS], 20K07774, 17K09052, 20K09259, 17K08970, and 17H04324) and Japan Agency for Medical Research and Development (AMED; no. JP19ek0610011).
ORCID iD: Kazutaka Ikeda https://orcid.org/0000-0001-8342-0278
Supplemental Material: Supplementary material for this article is available online.
References
- 1.Fukuda K, Hayashida M, Ide S, Saita N, Kokita Y, Kasai S, Nishizawa D, Ogai Y, Hasegawa J, Nagashima M, Tagami M, Komatsu H, Sora I, Koga H, Kaneko Y, Ikeda K. Association between OPRM1 gene polymorphisms and fentanyl sensitivity in patients undergoing painful cosmetic surgery. Pain 2009; 147: 194–201. [DOI] [PubMed] [Google Scholar]
- 2.Ikeda K, Ide S, Han W, Hayashida M, Uhl GR, Sora I. How individual sensitivity to opiates can be predicted by gene analyses. Trends Pharmacol Sci 2005; 26: 311–317. [DOI] [PubMed] [Google Scholar]
- 3.Mrozkova P, Palecek J, Spicarova D. The role of protease-activated receptor type 2 in nociceptive signaling and pain. Physiol Res 2016; 65: 357–367. [DOI] [PubMed] [Google Scholar]
- 4.Antoniak S, Mackman N. Multiple roles of the coagulation protease cascade during virus infection. Blood 2014; 123: 2605–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang M, Gao CX, Wang YP, Ma KT, Li L, Yin JW, Dai ZG, Wang S, Si JQ. The association between the expression of PAR2 and TMEM16A and neuropathic pain. Mol Med Rep 2018; 17: 3744–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park GH, Jeon SJ, Ryu JR, Choi MS, Han SH, Yang SI, Ryu JH, Cheong JH, Shin CY, Ko KH. Essential role of mitogen-activated protein kinase pathways in protease activated receptor 2-mediated nitric-oxide production from rat primary astrocytes. Nitric Oxide 2009; 21: 110–119. [DOI] [PubMed] [Google Scholar]
- 7.Bushell TJ, Cunningham MR, McIntosh KA, Moudio S, Plevin R. Protease-activated receptor 2: are common functions in glial and immune cells linked to inflammation-related CNS disorders? Curr Drug Targets 2016; 17: 1861–1870. [DOI] [PubMed] [Google Scholar]
- 8.Ito M, Ono K, Hitomi S, Nodai T, Sago T, Yamaguchi K, Harano N, Gunnjigake K, Hosokawa R, Kawamoto T, Inenaga K. Prostanoid-dependent spontaneous pain and PAR2-dependent mechanical allodynia following oral mucosal trauma: involvement of TRPV1, TRPA1 and TRPV4. Mol Pain 2017; 13: 1744806917704138–/1744806917704107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jimenez-Vargas NN, Pattison LA, Zhao P, Lieu T, Latorre R, Jensen DD, Castro J, Aurelio L, Le GT, Flynn B, Herenbrink CK, Yeatman HR, Edgington-Mitchell L, Porter CJH, Halls ML, Canals M, Veldhuis NA, Poole DP, McLean P, Hicks GA, Scheff N, Chen E, Bhattacharya A, Schmidt BL, Brierley SM, Vanner SJ, Bunnett NW. Protease-activated receptor-2 in endosomes signals persistent pain of irritable bowel syndrome. Proc Natl Acad Sci U S A 2018; 115: E7438–E7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao P, Lieu T, Barlow N, Metcalf M, Veldhuis NA, Jensen DD, Kocan M, Sostegni S, Haerteis S, Baraznenok V, Henderson I, Lindstrom E, Guerrero-Alba R, Valdez-Morales EE, Liedtke W, McIntyre P, Vanner SJ, Korbmacher C, Bunnett NW. Cathepsin S causes inflammatory pain via biased agonism of PAR2 and TRPV4. J Biol Chem 2014; 289: 27215–27234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao P, Lieu T, Barlow N, Sostegni S, Haerteis S, Korbmacher C, Liedtke W, Jimenez-Vargas NN, Vanner SJ, Bunnett NW. Neutrophil elastase activates protease-activated receptor-2 (PAR2) and transient receptor potential vanilloid 4 (TRPV4) to cause inflammation and pain. J Biol Chem 2015; 290: 13875–13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao Y, Hou W, Hua B. Protease-activated receptor 2 signalling pathways: a role in pain processing. Expert Opin Ther Targets 2014; 18: 15–27. [DOI] [PubMed] [Google Scholar]
- 13.Huang ZJ, Li HC, Cowan AA, Liu S, Zhang YK, Song XJ. Chronic compression or acute dissociation of dorsal root ganglion induces cAMP-dependent neuronal hyperexcitability through activation of PAR2. Pain 2012; 153: 1426–1437. [DOI] [PubMed] [Google Scholar]
- 14.Lam DK, Dang D, Zhang J, Dolan JC, Schmidt BL. Novel animal models of acute and chronic cancer pain: a pivotal role for PAR2. J Neurosci 2012; 32: 14178–14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramsey LB, Pounds S, Cheng C, Cao X, Yang W, Smith C, Karol SE, Liu C, Panetta JC, Inaba H, Rubnitz JE, Metzger ML, Ribeiro RC, Sandlund JT, Jeha S, Pui CH, Evans WE, Relling MV. Genetics of pleiotropic effects of dexamethasone. Pharmacogenet Genomics 2017; 27: 294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grundberg E, Adoue V, Kwan T, Ge B, Duan QL, Lam KC, Koka V, Kindmark A, Weiss ST, Tantisira K, Mallmin H, Raby BA, Nilsson O, Pastinen T. Global analysis of the impact of environmental perturbation on cis-regulation of gene expression. PLoS Genet 2011; 7: e1001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spicarova D, Nerandzic V, Palecek J. Update on the role of spinal cord TRPV1 receptors in pain modulation. Physiol Res 2014; 63: S225–S236. [DOI] [PubMed] [Google Scholar]
- 18.Spicarova D, Adamek P, Kalynovska N, Mrozkova P, Palecek J. TRPV1 receptor inhibition decreases CCL2-induced hyperalgesia. Neuropharmacology 2014; 81: 75–84. [DOI] [PubMed] [Google Scholar]
- 19.Uchytilova E, Spicarova D, Palecek J. TRPV1 antagonist attenuates postoperative hypersensitivity by central and peripheral mechanisms. Mol Pain 2014; 10: 67–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai Y. TRPs and pain. Semin Immunopathol 2016; 38: 277–291. [DOI] [PubMed] [Google Scholar]
- 21.Moore C, Gupta R, Jordt SE, Chen Y, Liedtke WB. Regulation of pain and itch by TRP channels. Neurosci Bull 2018; 34: 120–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weyer AD, Lehto SG. Development of TRPM8 antagonists to treat chronic pain and migraine. Pharmaceuticals (Basel ) 2017; 10: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takaishi M, Uchida K, Suzuki Y, Matsui H, Shimada T, Fujita F, Tominaga M. Reciprocal effects of capsaicin and menthol on thermosensation through regulated activities of TRPV1 and TRPM8. J Physiol Sci 2016; 66: 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L, Rohacs T. Regulation of the cold-sensing TRPM8 channels by phosphoinositides and Gq-coupled receptors. Channels (Austin) 2020; 14: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bisgaard T, Klarskov B, Rosenberg J, Kehlet H. Characteristics and prediction of early pain after laparoscopic cholecystectomy. Pain 2001; 90: 261–269. [DOI] [PubMed] [Google Scholar]
- 26.Martikainen IK, Narhi MV, Pertovaara A. Spatial integration of cold pressor pain sensation in humans. Neurosci Lett 2004; 361: 140–143. [DOI] [PubMed] [Google Scholar]
- 27.Nishizawa D, Fukuda K, Kasai S, Hasegawa J, Aoki Y, Nishi A, Saita N, Koukita Y, Nagashima M, Katoh R, Satoh Y, Tagami M, Higuchi S, Ujike H, Ozaki N, Inada T, Iwata N, Sora I, Iyo M, Kondo N, Won MJ, Naruse N, Uehara-Aoyama K, Itokawa M, Koga M, Arinami T, Kaneko Y, Hayashida M, Ikeda K. Genome-wide association study identifies a potent locus associated with human opioid sensitivity. Mol Psychiatry 2014; 19: 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muraoka W, Nishizawa D, Fukuda K, Kasai S, Hasegawa J, Wajima K, Nakagawa T, Ikeda K. Association between UGT2B7 gene polymorphisms and fentanyl sensitivity in patients undergoing painful orthognathic surgery. Mol Pain 2016; 12: 1744806916683182–1744806916683101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amano K, Nishizawa D, Mieda T, Tsujita M, Kitamura A, Hasegawa J, Inada E, Hayashida M, Ikeda K. Opposite associations between the rs3845446 single-nucleotide polymorphism of the CACNA1E gene and postoperative pain-related phenotypes in gastrointestinal surgery versus previously reported orthognathic surgery. J Pain 2016; 17: 1126–1134. [DOI] [PubMed] [Google Scholar]
- 30.Kayama Y, Shibata M, Takizawa T, Ibata K, Nakahara J, Shimizu T, Toriumi H, Yuzaki M, Suzuki N. Signaling pathways relevant to nerve growth factor-induced upregulation of transient receptor potential M8 expression. Neuroscience 2017; 367: 178–188. [DOI] [PubMed] [Google Scholar]
- 31.Obata K, Katsura H, Mizushima T, Yamanaka H, Kobayashi K, Dai Y, Fukuoka T, Tokunaga A, Tominaga M, Noguchi K. TRPA1 induced in sensory neurons contributes to cold hyperalgesia after inflammation and nerve injury. J Clin Invest 2005; 115: 2393–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Mikrani R, He Y, Faran Ashraf Baig MM, Abbas M, Naveed M, Tang M, Zhang Q, Li C, Zhou X. TRPM8 channels: a review of distribution and clinical role. Eur J Pharmacol 2020; 882: 173312–173307. [DOI] [PubMed] [Google Scholar]
- 33.Liu L, Yudin Y, Nagwekar J, Kang C, Shirokova N, Rohacs T. G alphaq sensitizes TRPM8 to inhibition by PI(4,5)P2 depletion upon receptor activation. J Neurosci 2019; 39: 6067–6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung SR, Seo JB, Deng Y, Asbury CL, Hille B, Koh DS. Contributions of protein kinases and beta-arrestin to termination of protease-activated receptor 2 signaling. J Gen Physiol 2016; 147: 255–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.del Camino D, Murphy S, Heiry M, Barrett LB, Earley TJ, Cook CA, Petrus MJ, Zhao M, D'Amours M, Deering N, Brenner GJ, Costigan M, Hayward NJ, Chong JA, Fanger CM, Woolf CJ, Patapoutian A, Moran MM. TRPA1 contributes to cold hypersensitivity. J Neurosci 2010; 30: 15165–15174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKemy DD. Chapter 13 TRPM8: the cold and menthol receptor. In: Liedtke WB, Heller S. (eds) TRP ion channel function in sensory transduction and cellular signaling cascades. 1st ed. Boca Raton: Taylor and Francis Group, 2006, pp.596–606. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-mpx-10.1177_17448069211002009 for Cold pain sensitivity is associated with single-nucleotide polymorphisms of PAR2/F2RL1 and TRPM8 by Moe Soeda, Seii Ohka, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Tatsuya Ichinohe, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain
Supplemental material, sj-xlsx-2-mpx-10.1177_17448069211002009 for Cold pain sensitivity is associated with single-nucleotide polymorphisms of PAR2/F2RL1 and TRPM8 by Moe Soeda, Seii Ohka, Daisuke Nishizawa, Junko Hasegawa, Kyoko Nakayama, Yuko Ebata, Tatsuya Ichinohe, Ken-ichi Fukuda and Kazutaka Ikeda in Molecular Pain